2JYD
 
 | | Structure of the fifth zinc finger of Myelin Transcription Factor 1 | | Descriptor: | F5 domain of Myelin transcription factor 1, ZINC ION | | Authors: | Gamsjaeger, R, Swanton, M.K, Kobus, F.J, Lehtomaki, E, Lowry, J.A, Kwan, A.H, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2007-12-12 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and biophysical analysis of the DNA binding properties of myelin transcription factor 1.
J.Biol.Chem., 283, 2008
|
|
2XU7
 
 | | Structural basis for RbAp48 binding to FOG-1 | | Descriptor: | HISTONE-BINDING PROTEIN RBBP4, TETRAETHYLENE GLYCOL, ZINC FINGER PROTEIN ZFPM1 | | Authors: | Lejon, S, Thong, S.Y, Murthy, A, Blobel, G.A, Mackay, J.P, Murzina, N.V, Laue, E.D. | | Deposit date: | 2010-10-15 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights Into Association of the Nurd Complex with Fog-1 from the Crystal Structure of an Rbap48-Fog- 1 Complex.
J.Biol.Chem., 286, 2011
|
|
1FV5
 
 | | SOLUTION STRUCTURE OF THE FIRST ZINC FINGER FROM THE DROSOPHILA U-SHAPED TRANSCRIPTION FACTOR | | Descriptor: | FIRST ZINC FINGER OF U-SHAPED, ZINC ION | | Authors: | Liew, C.K, Kowalski, K, Fox, A.H, Newton, A, Sharpe, B.K, Crossley, M, Mackay, J.P. | | Deposit date: | 2000-09-18 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions.
Structure Fold.Des., 8, 2000
|
|
1FU9
 
 | | SOLUTION STRUCTURE OF THE NINTH ZINC-FINGER DOMAIN OF THE U-SHAPED TRANSCRIPTION FACTOR | | Descriptor: | U-SHAPED TRANSCRIPTIONAL COFACTOR, ZINC ION | | Authors: | Liew, C.K, Kowalski, K, Fox, A.H, Newton, A, Sharpe, B.K, Crossley, M, Mackay, J.P. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions.
Structure Fold.Des., 8, 2000
|
|
1GNF
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZINC FINGER OF MURINE GATA-1, NMR, 25 STRUCTURES | | Descriptor: | TRANSCRIPTION FACTOR GATA-1, ZINC ION | | Authors: | Kowalski, K, Czolij, R, King, G.F, Crossley, M, Mackay, J.P. | | Deposit date: | 1998-10-12 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal zinc finger of GATA-1 reveals a specific binding face for the transcriptional co-factor FOG.
J.Biomol.NMR, 13, 1999
|
|
1J5B
 
 | | Solution structure of a hydrophobic analogue of the winter flounder antifreeze protein | | Descriptor: | Antifreeze protein type 1 analogue | | Authors: | Liepinsh, E, Otting, G, Harding, M.M, Ward, L.G, Mackay, J.P, Haymet, A.D. | | Deposit date: | 2002-03-22 | | Release date: | 2002-03-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a hydrophobic analogue of the winter flounder antifreeze protein.
Eur.J.Biochem., 269, 2002
|
|
1K2F
 
 | | siah, Seven In Absentia Homolog | | Descriptor: | BETA-MERCAPTOETHANOL, ZINC ION, siah-1A protein | | Authors: | Polekhina, G, House, C.M, Traficante, N, Mackay, J.P, Relaix, F, Sassoon, D.A, Parker, M.W, Bowtell, D.D.L. | | Deposit date: | 2001-09-26 | | Release date: | 2001-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling.
Nat.Struct.Biol., 9, 2002
|
|
1JN7
 
 | |
1MM3
 
 | | Solution structure of the 2nd PHD domain from Mi2b with C-terminal loop replaced by corresponding loop from WSTF | | Descriptor: | Mi2-beta(Chromodomain helicase-DNA-binding protein 4) and transcription factor WSTF, ZINC ION | | Authors: | Kwan, A.H.Y, Gell, D.A, Verger, A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2002-09-02 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Engineering a Protein Scaffold from a PHD Finger
structure, 11, 2003
|
|
1M36
 
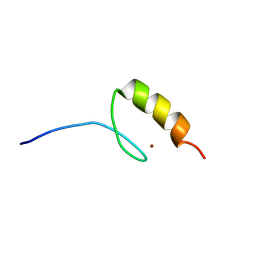 | |
1LIQ
 
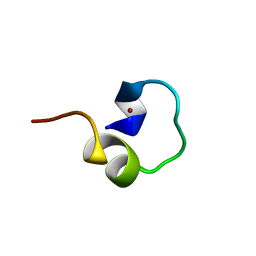 | | Non-native Solution Structure of a fragment of the CH1 domain of CBP | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Matthews, J.M, Kwan, A.H.Y, Newton, A, Gell, D.A, Crossley, M, Mackay, J.P. | | Deposit date: | 2002-04-18 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Zinc Binding Fold Underlines the Versatility of Zinc Binding Modules in Protein Evolution
Structure, 10, 2002
|
|
1P7A
 
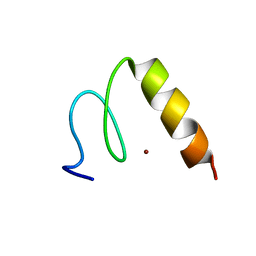 | | Solution Structure of the Third Zinc Finger from BKLF | | Descriptor: | Kruppel-like factor 3, ZINC ION | | Authors: | Simpson, R.J.Y, Cram, E.D, Czolij, R, Matthews, J.M, Crossley, M, Mackay, J.P. | | Deposit date: | 2003-04-30 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | CCHX zinc finger derivatives retain the ability to bind Zn(II) and mediate protein-DNA interactions.
J.Biol.Chem., 278, 2003
|
|
1MM2
 
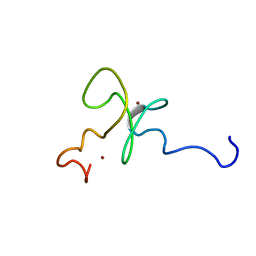 | | Solution structure of the 2nd PHD domain from Mi2b | | Descriptor: | Mi2-beta, ZINC ION | | Authors: | Kwan, A.H.Y, Gell, D.A, Verger, A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2002-09-02 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Engineering a Protein Scaffold from a PHD Finger
structure, 11, 2003
|
|
1N0Z
 
 | | Solution structure of the first zinc-finger domain from ZNF265 | | Descriptor: | ZINC ION, ZNF265 | | Authors: | Plambeck, C.A, Fairley, K, Kwan, A.H.Y, Westman, B.J, Adams, D, Morris, B, Mackay, J.P. | | Deposit date: | 2002-10-15 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the zinc finger domain from human splicing factor ZNF265 fold
J.BIOL.CHEM., 278, 2003
|
|
1PM4
 
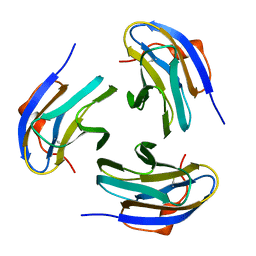 | | Crystal structure of Yersinia pseudotuberculosis-derived mitogen (YPM) | | Descriptor: | YPM | | Authors: | Donadini, R, Liew, C.W, Kwan, A.H, Mackay, J.P, Fields, B.A. | | Deposit date: | 2003-06-09 | | Release date: | 2004-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Crystal and Solution Structures of a Superantigen from Yersinia pseudotuberculosis Reveal a Jelly-Roll Fold.
Structure, 12, 2004
|
|
1POQ
 
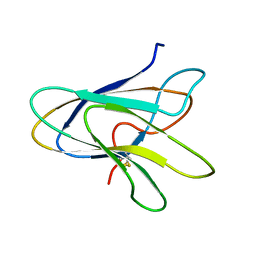 | | Solution Structure of a Superantigen from Yersinia pseudotuberculosis | | Descriptor: | YPM | | Authors: | Donadini, R, Liew, C.W, Kwan, A.H, Mackay, J.P, Fields, B.A. | | Deposit date: | 2003-06-16 | | Release date: | 2004-01-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Crystal and Solution Structures of a Superantigen from Yersinia pseudotuberculosis Reveal a Jelly-Roll Fold.
Structure, 12, 2004
|
|
1SRK
 
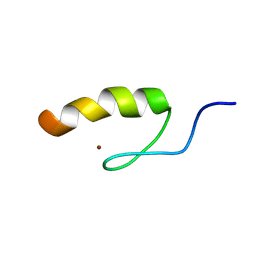 | | Solution structure of the third zinc finger domain of FOG-1 | | Descriptor: | ZINC ION, Zinc finger protein ZFPM1 | | Authors: | Simpson, R.J.Y, Lee, S.H.Y, Bartle, N, Matthews, J.M, Mackay, J.P, Crossley, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Classic Zinc Finger from Friend of GATA Mediates an Interaction with the Coiled-coil of Transforming Acidic Coiled-coil 3.
J.Biol.Chem., 279, 2004
|
|
1U85
 
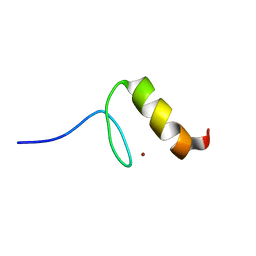 | |
1U86
 
 | |
1WO3
 
 | | Solution structure of Minimal Mutant 1 (MM1): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO6
 
 | | Solution structure of Designed Functional Finger 5 (DFF5): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO4
 
 | | Solution structure of Minimal Mutant 2 (MM2): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO5
 
 | | Solution structure of Designed Functional Finger 2 (DFF2): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO7
 
 | | Solution structure of Designed Functional Finger 7 (DFF7): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1XZY
 
 | | Solution structure of the P30-trans form of Alpha Hemoglobin Stabilizing Protein (AHSP) | | Descriptor: | Alpha-hemoglobin stabilizing protein | | Authors: | Gell, D.A, Feng, L, Zhou, S, Kong, Y, Lee, C, Weiss, M.J, Shi, Y, Mackay, J.P. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular mechanism of AHSP-mediated stabilization of alpha-hemoglobin
Cell(Cambridge,Mass.), 119, 2004
|
|
