1QVT
 
 | |
1QX7
 
 | |
1RZR
 
 | | crystal structure of transcriptional regulator-phosphoprotein-DNA complex | | Descriptor: | 5'-D(*CP*TP*GP*AP*AP*AP*GP*CP*GP*CP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*AP*GP*CP*GP*CP*TP*TP*TP*CP*AP*G)-3', Glucose-resistance amylase regulator, ... | | Authors: | Schumacher, M.A, Allen, G.S, Brennan, R.G. | | Deposit date: | 2003-12-27 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P.
Cell(Cambridge,Mass.), 118, 2004
|
|
1SDK
 
 | | CROSS-LINKED, CARBONMONOXY HEMOGLOBIN A | | Descriptor: | 1,3,5-BENZENETRICARBOXYLIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-02-26 | | Release date: | 1996-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric transition intermediates modelled by crosslinked haemoglobins.
Nature, 375, 1995
|
|
4PQL
 
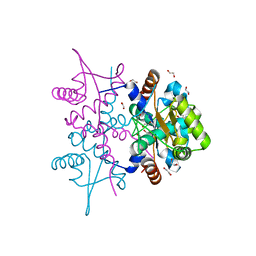 | | N-Terminal domain of DNA binding protein | | Descriptor: | 1,2-ETHANEDIOL, Truncated replication protein RepA | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N.K. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OB4
 
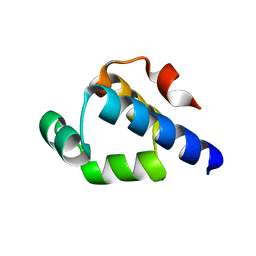 | | Structure of the S. venezulae BldD DNA-binding domain | | Descriptor: | Putative DNA-binding protein | | Authors: | schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
4PT7
 
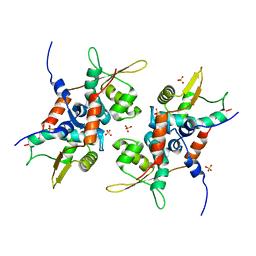 | | Structure of initiator | | Descriptor: | Replication initiator A family protein, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PTA
 
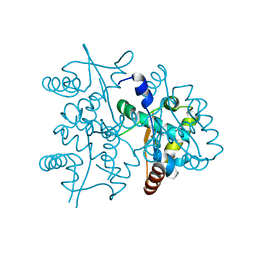 | | Structure of MDR initiator | | Descriptor: | Replication initiator protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6003 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1SDL
 
 | | CROSS-LINKED, CARBONMONOXY HEMOGLOBIN A | | Descriptor: | 1,3,5-BENZENETRICARBOXYLIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-02-26 | | Release date: | 1996-08-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric transition intermediates modelled by crosslinked haemoglobins.
Nature, 375, 1995
|
|
1SXH
 
 | | apo structure of B. megaterium transcription regulator | | Descriptor: | Glucose-resistance amylase regulator | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
1SXI
 
 | | Structure of apo transcription regulator B. megaterium | | Descriptor: | Glucose-resistance amylase regulator, MAGNESIUM ION | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
1SXG
 
 | | Structural studies on the apo transcription factor form B. megaterium | | Descriptor: | 2-PHENYLAMINO-ETHANESULFONIC ACID, Glucose-resistance amylase regulator | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
4PQK
 
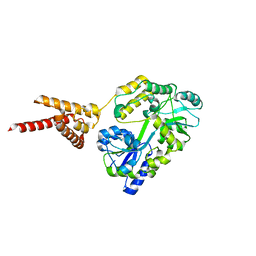 | | C-Terminal domain of DNA binding protein | | Descriptor: | Maltose ABC transporter periplasmic protein, Truncated replication protein RepA, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N.K. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1PP8
 
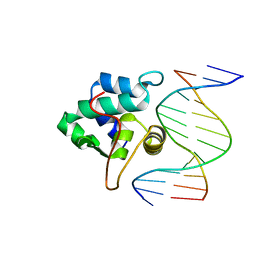 | | crystal structure of the T. vaginalis IBP39 Initiator binding domain (IBD) bound to the alpha-SCS Inr element | | Descriptor: | 39 kDa initiator binding protein, ALPHA-SCS INR, SULFATE ION | | Authors: | Schumacher, M.A, Lau, A.O.T, Johnson, P.J. | | Deposit date: | 2003-06-16 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Basis of Core Promoter Recognition in a Primitive Eukaryote
Cell(Cambridge,Mass.), 115, 2003
|
|
4OAZ
 
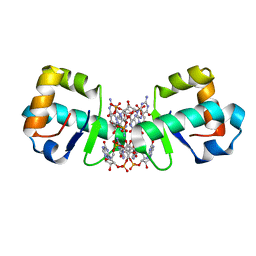 | | BldD CTD-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative DNA-binding protein | | Authors: | Schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R.G. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
1JJX
 
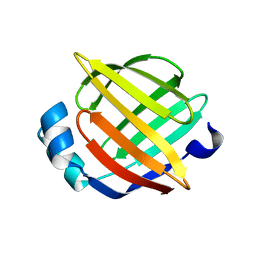 | | Solution Structure of Recombinant Human Brain-type Fatty acid Binding Protein | | Descriptor: | BRAIN-TYPE FATTY ACID BINDING PROTEIN | | Authors: | Rademacher, M, Zimmerman, A.W, Rueterjans, H, Veerkamp, J.H, Luecke, C. | | Deposit date: | 2001-07-10 | | Release date: | 2002-10-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of fatty acid-binding protein from human brain.
Mol.Cell.Biochem., 239, 2002
|
|
1PP7
 
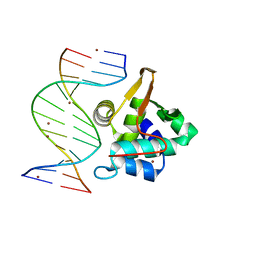 | | Crystal structure of the T. vaginalis Initiator binding protein bound to the ferredoxin Inr | | Descriptor: | 39 kDa initiator binding protein, FERREDOXIN INR, ZINC ION | | Authors: | Schumacher, M.A, Lau, A.O.T, Johnson, P.J. | | Deposit date: | 2003-06-16 | | Release date: | 2003-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Basis of Core Promoter Recognition in a Primitive Eukaryote
Cell(Cambridge,Mass.), 115, 2003
|
|
1Q87
 
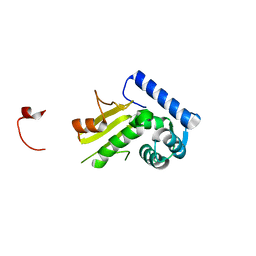 | |
1PNR
 
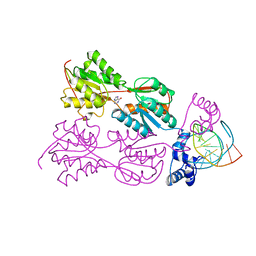 | | PURINE REPRESSOR-HYPOXANTHINE-PURF-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*TP*TP*TP*TP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1995-03-29 | | Release date: | 1995-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices.
Science, 266, 1994
|
|
1Q89
 
 | |
1Q88
 
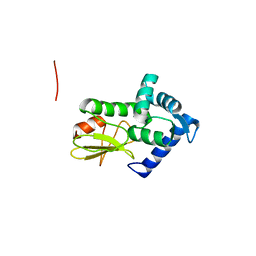 | |
7NWW
 
 | | CspA-27 cotranslational folding intermediate 1 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-03-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OIF
 
 | | CspA-27 cotranslational folding intermediate 2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OIG
 
 | | CspA-27 cotranslational folding intermediate 3 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OT5
 
 | | CspA-70 cotranslational folding intermediate 1 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-06-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
