3KIA
 
 | | Crystal structure of mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Macedo-Ribeiro, S, Pereira, P.J.B, Empadinhas, N, da Costa, M.S. | | Deposit date: | 2009-11-01 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional and structural characterization of a novel mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus reveals its dual substrate specificity
Mol.Microbiol., 79, 2011
|
|
3O3P
 
 | | Crystal structure of R. xylanophilus MpgS in complex with GDP-Mannose | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Macedo-Ribeiro, S, Pereira, P.J.B, Empadinhas, N, da Costa, M.S. | | Deposit date: | 2010-07-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.529 Å) | | Cite: | Functional and structural characterization of a novel mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus reveals its dual substrate specificity
Mol.Microbiol., 79, 2011
|
|
2ODY
 
 | | Thrombin-bound boophilin displays a functional and accessible reactive-site loop | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Boophilin, PHOSPHATE ION, ... | | Authors: | Macedo-Ribeiro, S, Fuentes-Prior, P, Pereira, P.J.B. | | Deposit date: | 2006-12-27 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Isolation, cloning and structural characterisation of boophilin, a multifunctional kunitz-type proteinase inhibitor from the cattle tick.
Plos One, 3, 2008
|
|
3F1Y
 
 | | Mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mannosyl-3-phosphoglycerate synthase | | Authors: | Macedo-Ribeiro, S, Pereira, P.J.B, Empadinhas, N, da Costa, M.S. | | Deposit date: | 2008-10-28 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural characterization of a novel mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus reveals its dual substrate specificity
Mol.Microbiol., 79, 2011
|
|
5TNZ
 
 | | HtrA2 S142D mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SODIUM ION, Serine protease HTRA2, ... | | Authors: | Macedo-Ribeiro, S, Merski, M, Pereira, P.J.B. | | Deposit date: | 2016-10-15 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular motion regulates the activity of the Mitochondrial Serine Protease HtrA2.
Cell Death Dis, 8, 2017
|
|
1H98
 
 | | New Insights into Thermostability of Bacterial Ferredoxins: High Resolution Crystal Structure of the Seven-Iron Ferredoxin from Thermus thermophilus | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Macedo-Ribeiro, S, Martins, B.M, Pereira, P.J.B, Buse, G, Huber, R, Soulimane, T. | | Deposit date: | 2001-03-05 | | Release date: | 2001-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | New Insights Into the Thermostability of Bacterial Ferredoxins: High-Resolution Crystal Structure of the Seven-Iron Ferredoxin from Thermus Thermophilus
J.Biol.Inorg.Chem., 6, 2001
|
|
1VJW
 
 | | STRUCTURE OF OXIDOREDUCTASE (NADP+(A),FERREDOXIN(A)) | | Descriptor: | FERREDOXIN(A), IRON/SULFUR CLUSTER | | Authors: | Macedo-Ribeiro, S, Darimont, B, Sterner, R, Huber, R. | | Deposit date: | 1996-10-09 | | Release date: | 1996-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small structural changes account for the high thermostability of 1[4Fe-4S] ferredoxin from the hyperthermophilic bacterium Thermotoga maritima.
Structure, 4, 1996
|
|
1VNE
 
 | |
1VNF
 
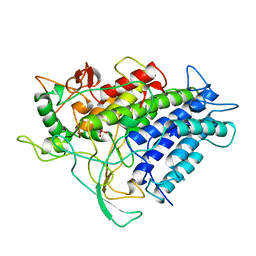 | |
1VNG
 
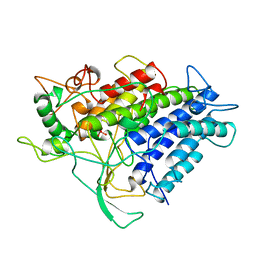 | |
1VNH
 
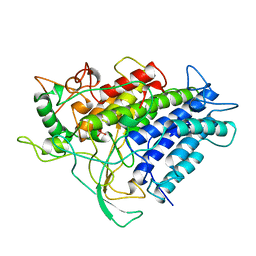 | |
1VNI
 
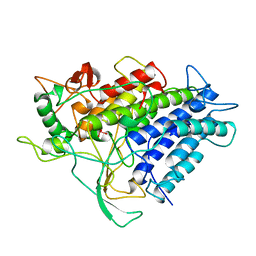 | |
1CZV
 
 | | CRYSTAL STRUCTURE OF THE C2 DOMAIN OF HUMAN COAGULATION FACTOR V: DIMERIC CRYSTAL FORM | | Descriptor: | PROTEIN (COAGULATION FACTOR V) | | Authors: | Macedo-Ribeiro, S, Bode, W, Huber, R, Kane, W.H, Fuentes-Prior, P. | | Deposit date: | 1999-09-07 | | Release date: | 1999-11-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the membrane-binding C2 domain of human coagulation factor V.
Nature, 402, 1999
|
|
1CZS
 
 | | CRYSTAL STRUCTURE OF THE C2 DOMAIN OF HUMAN COAGULATION FACTOR V: COMPLEX WITH PHENYLMERCURY | | Descriptor: | PHENYLMERCURY, PROTEIN (COAGULATION FACTOR V) | | Authors: | Macedo-Ribeiro, S, Bode, W, Huber, R, Kane, W.H, Fuentes-Prior, P. | | Deposit date: | 1999-09-07 | | Release date: | 1999-11-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the membrane-binding C2 domain of human coagulation factor V.
Nature, 402, 1999
|
|
1CZT
 
 | | CRYSTAL STRUCTURE OF THE C2 DOMAIN OF HUMAN COAGULATION FACTOR V | | Descriptor: | PROTEIN (COAGULATION FACTOR V) | | Authors: | Macedo-Ribeiro, S, Bode, W, Huber, R, Kane, W.H, Fuentes-Prior, P. | | Deposit date: | 1999-09-07 | | Release date: | 1999-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of the membrane-binding C2 domain of human coagulation factor V.
Nature, 402, 1999
|
|
1VNS
 
 | |
7QCP
 
 | |
7QI9
 
 | |
7QIB
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UDP | | Descriptor: | CHLORIDE ION, Glucosyl-3-phosphoglycerate synthase, MAGNESIUM ION, ... | | Authors: | Silva, A, Nunes-Costa, D, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UDP
To Be Published
|
|
7QOQ
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UMP and Magnesium | | Descriptor: | CHLORIDE ION, Glucosyl-3-phosphoglycerate synthase, MAGNESIUM ION, ... | | Authors: | Silva, A, Barbosa Pereira, P.J, Macedo-Ribeiro, S, Costa, D. | | Deposit date: | 2021-12-27 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UMP and Magnesium
To Be Published
|
|
7NZZ
 
 | |
4WZN
 
 | | CRYSTAL STRUCTURE OF THE 2B PROTEIN SOLUBLE DOMAIN FROM HEPATITIS A VIRUS | | Descriptor: | GLYCEROL, Genome polyprotein | | Authors: | Garriga, D, Vives-Adrian, L, Buxaderas, M, Ferreira-da-Silva, F, Almeida, B, Macedo-Ribeiro, S, Pereira, P.J, Verdaguer, N. | | Deposit date: | 2014-11-20 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Host Membrane Remodeling Induced by Protein 2B of Hepatitis A Virus.
J.Virol., 89, 2015
|
|
6Q5T
 
 | |
3BB0
 
 | |
1A0L
 
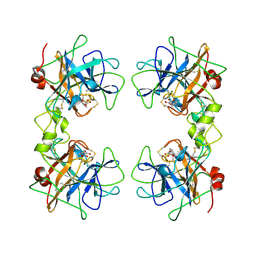 | | HUMAN BETA-TRYPTASE: A RING-LIKE TETRAMER WITH ACTIVE SITES FACING A CENTRAL PORE | | Descriptor: | (2S)-3-(4-carbamimidoylphenyl)-2-hydroxypropanoic acid, BETA-TRYPTASE | | Authors: | Pereira, P.J.B, Bergner, A, Macedo-Ribeiro, S, Huber, R, Matschiner, G, Fritz, H, Sommerhoff, C.P, Bode, W. | | Deposit date: | 1997-12-03 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human beta-tryptase is a ring-like tetramer with active sites facing a central pore.
Nature, 392, 1998
|
|
