3T35
 
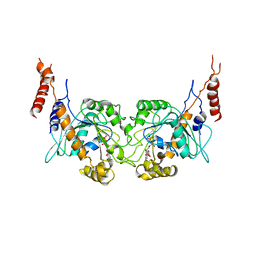 | | Arabidopsis thaliana dynamin-related protein 1A in postfission state | | Descriptor: | Dynamin-related protein 1A, LINKER, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yan, L.M, Ma, Y.Y, Sun, Y.N, Lou, Z.Y. | | Deposit date: | 2011-07-24 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.592 Å) | | Cite: | Structural basis for mechanochemical role of Arabidopsis thaliana dynamin-related protein in membrane fission
J Mol Cell Biol, 3, 2011
|
|
3T34
 
 | | Arabidopsis thaliana dynamin-related protein 1A (AtDRP1A) in prefission state | | Descriptor: | Dynamin-related protein 1A, LINKER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yan, L.M, Ma, Y.Y, Sun, Y.N, Lou, Z.Y. | | Deposit date: | 2011-07-24 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structural basis for mechanochemical role of Arabidopsis thaliana dynamin-related protein in membrane fission
J Mol Cell Biol, 3, 2011
|
|
7VZF
 
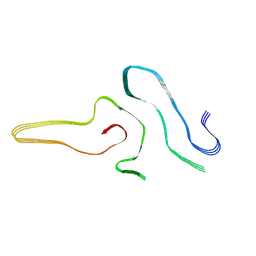 | | Cryo-EM structure of amyloid fibril formed by full-length human SOD1 | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, L.Q, Ma, Y.Y, Yuan, H.Y, Zhao, K, Zhang, M.Y, Wang, Q, Huang, X, Xu, W.C, Chen, J, Li, D, Zhang, D.L, Zou, L.Y, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human SOD1 reveals its conformational conversion.
Nat Commun, 13, 2022
|
|
3UIM
 
 | |
7U3P
 
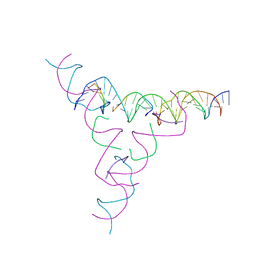 | | [3T7] Self-assembling tensegrity triangle with three turns of DNA per axis with R3 symmetry | | Descriptor: | DNA (31-MER), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), DNA (5'-D(P*GP*GP*CP*TP*GP*C)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Lu, B, Ma, Y, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.06 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
6K20
 
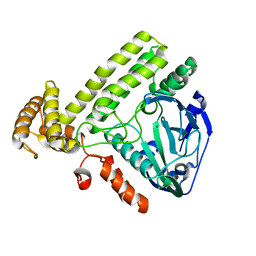 | | Structure of Apo YdiU | | Descriptor: | MAGNESIUM ION, Protein adenylyltransferase SelO | | Authors: | Li, B, Yang, Y, Ma, Y. | | Deposit date: | 2019-05-13 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of Apo-YdiU
To Be Published
|
|
7U3Q
 
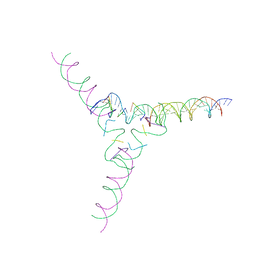 | | [4T7] Self-assembling tensegrity triangle with four turns of DNA per axis with R3 symmetry | | Descriptor: | DNA (29-MER), DNA (42-MER), DNA (5'-D(P*AP*CP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Lu, B, Ma, Y, Seeman, N.C, Sha, R, Ohayon, Y.P, Huang, Q. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (9.32 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
3VSU
 
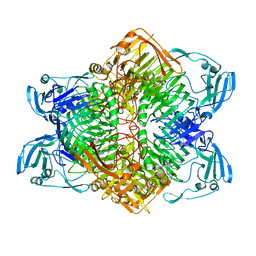 | | The complex structure of XylC with xylobiose | | Descriptor: | Xylosidase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
3VST
 
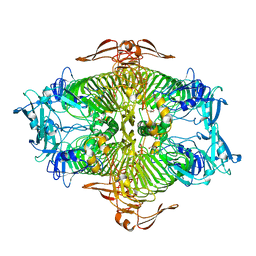 | | The complex structure of XylC with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Xylosidase | | Authors: | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
5HT6
 
 | | Crystal structure of the SET domain of the human MLL5 methyltransferase | | Descriptor: | Histone-lysine N-methyltransferase 2E | | Authors: | le Maire, A, Mas-y-Mas, S, Dumas, C, Lebedev, A. | | Deposit date: | 2016-01-26 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | The Human Mixed Lineage Leukemia 5 (MLL5), a Sequentially and Structurally Divergent SET Domain-Containing Protein with No Intrinsic Catalytic Activity.
Plos One, 11, 2016
|
|
