7GZ3
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000090-001 | | Descriptor: | (2S,3S)-N-(5,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-2-methyloxolane-3-carboxamide, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZK
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000877-001 | | Descriptor: | N-{(1R)-1-[(3R)-oxolan-3-yl]ethyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.163 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZF
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000605-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-[(1-methyl-1H-pyrazolo[3,4-b]pyridine-4-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H02
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000455-001 | | Descriptor: | N-[(1S)-1-(3,4-dihydro-2H-1lambda~4~-thiophen-5-yl)-2-methylpropyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7DKN
 
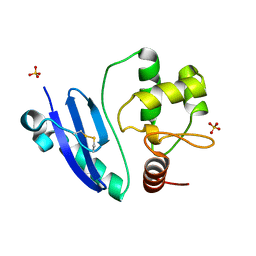 | | Crystal structure of Sulfurisphaera tokodaii O6-methylguanine methyltransferase | | Descriptor: | Methylated-DNA--protein-cysteine methyltransferase, SULFATE ION | | Authors: | Kikuchi, M, Yamauchi, T, Iizuka, Y, Tsunoda, M. | | Deposit date: | 2020-11-25 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7GZY
 
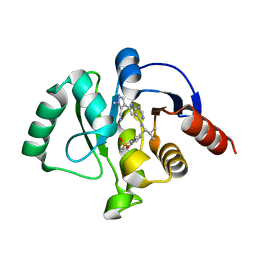 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011144-001 | | Descriptor: | (4M)-4-(4-{[(1S)-1-(1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~-benzothiopyran-7-yl)-2-methylpropyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-1-methyl-1H-pyrazole-5-carbonitrile, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H0K
 
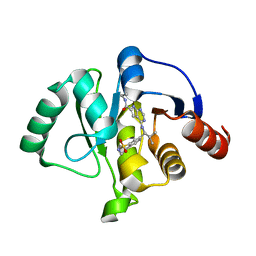 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011076-001 | | Descriptor: | 4-{[(1S)-1-(1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazin-7-yl)-2-methylpropyl]amino}-N-ethyl-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
6EHF
 
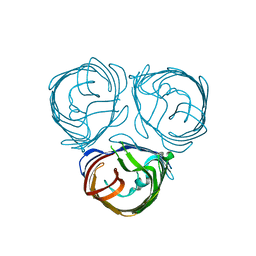 | |
7H0L
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011436-001 | | Descriptor: | 4-{[(S)-cyclopropyl(1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~-benzothiopyran-7-yl)methyl]amino}-N-ethyl-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H0T
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013772-001 | | Descriptor: | 7-[(1S)-1-{[(6M)-6-{3-[(4-acetylpiperazin-1-yl)methyl]phenyl}-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}-2-methylpropyl]-3,4-dihydro-1lambda~6~-benzothiopyran-1,1(2H)-dione, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H11
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013392-001 | | Descriptor: | 7-[(1R)-1-{[(6M)-6-(2,5-dihydrofuran-3-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}-2-methylpropyl]-3,4-dihydro-1lambda~6~-benzothiopyran-1,1(2H)-dione, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H15
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013387-001 | | Descriptor: | 7-[(1R)-1-{[(6M)-6-{2-[2-(dimethylamino)ethoxy]pyridin-4-yl}-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}-2-methylpropyl]-3,4-dihydro-1lambda~6~-benzothiopyran-1,1(2H)-dione, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H19
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013827-001 | | Descriptor: | 4-{[(1S)-1-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methylpropyl]amino}-N-ethyl-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
6RI3
 
 | | Dodecin from Streptomyces davaonensis | | Descriptor: | dodecin | | Authors: | Paithankar, K.S, Bourdeaux, F, Grininger, M, Ludwig, P, Mack, M. | | Deposit date: | 2019-04-23 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparative biochemical and structural analysis of the flavin-binding dodecins from Streptomyces davaonensis and Streptomyces coelicolor reveals striking differences with regard to multimerization.
Microbiology (Reading, Engl.), 165, 2019
|
|
1WT7
 
 | | Solution structure of BuTX-MTX: a butantoxin-maurotoxin chimera | | Descriptor: | BuTX-MTX | | Authors: | M'Barek, S, Chagot, B, Andreotti, N, Visan, V, Mansuelle, P, Grissmer, S, Marrakchi, M, El Ayeb, M, Sampieri, F, Darbon, H, Fajloun, Z, De Waard, M, Sabatier, J.-M. | | Deposit date: | 2004-11-16 | | Release date: | 2004-11-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Increasing the molecular contacts between maurotoxin and Kv1.2 channel augments ligand affinity.
Proteins, 60, 2005
|
|
4KDD
 
 | | Structure of Mycobacterium tuberculosis ribosome recycling factor in presence of detergent | | Descriptor: | CADMIUM ION, DECYL-BETA-D-MALTOPYRANOSIDE, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-24 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
8BTM
 
 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GdmF | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional studies of geldanamycin amide synthase ShGdmF
To Be Published
|
|
7GZB
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000495-001 | | Descriptor: | (2R)-(2,3-dihydro-1-benzofuran-5-yl)[(7H-pyrrolo[2,3-d]pyrimidine-4-carbonyl)amino]acetic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
1J0D
 
 | | ACC deaminase mutant complexed with ACC | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
7DK2
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW07 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW07 heavy chain, MW07 light chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M, Chen, S. | | Deposit date: | 2020-11-22 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Architectural versatility of spike neutralization by a SARS-CoV-2 antibody
To Be Published
|
|
7GZT
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0008348-001 | | Descriptor: | 4-{[(1S)-1-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methylpropyl]amino}-N-methyl-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
1WXU
 
 | | Solution structure of the SH3 domain of mouse peroxisomal biogenesis factor 13 | | Descriptor: | peroxisomal biogenesis factor 13 | | Authors: | Yoneyama, M, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-01 | | Release date: | 2005-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of mouse peroxisomal biogenesis factor 13
To be Published
|
|
7H00
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011221-001 | | Descriptor: | (4M)-4-(4-{[(1S)-1-(2,3-dihydro[1,4]dioxino[2,3-b]pyridin-6-yl)-2,2-dimethylpropyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-1-methyl-1H-pyrazole-5-carbonitrile, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
1SUH
 
 | | AMINO-TERMINAL DOMAIN OF EPITHELIAL CADHERIN IN THE CALCIUM BOUND STATE, NMR, 20 STRUCTURES | | Descriptor: | EPITHELIAL CADHERIN | | Authors: | Overduin, M, Tong, K.I, Kay, C.M, Ikura, M. | | Deposit date: | 1996-01-30 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C resonance assignments and monomeric structure of the amino-terminal extracellular domain of epithelial cadherin.
J.Biomol.NMR, 7, 1996
|
|
7GZ5
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000050-002 | | Descriptor: | 4-[4-(4-methylpyrimidin-2-yl)piperidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
