3NK3
 
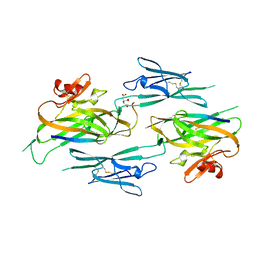 | | Crystal structure of full-length sperm receptor ZP3 at 2.6 A resolution | | Descriptor: | CITRATE ANION, Zona pellucida 3, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Monne, M, Jovine, L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into Egg Coat Assembly and Egg-Sperm Interaction from the X-Ray Structure of Full-Length ZP3.
Cell(Cambridge,Mass.), 143, 2010
|
|
1G66
 
 | | ACETYLXYLAN ESTERASE AT 0.90 ANGSTROM RESOLUTION | | Descriptor: | ACETYL XYLAN ESTERASE II, GLYCEROL, SULFATE ION | | Authors: | Ghosh, D, Sawicki, M, Lala, P, Erman, M, Pangborn, W, Eyzaguirre, J, Gutierrez, R, Jornvall, H, Thiel, D.J. | | Deposit date: | 2000-11-03 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Multiple conformations of catalytic serine and histidine in acetylxylan esterase at 0.90 A.
J.Biol.Chem., 276, 2001
|
|
1YF5
 
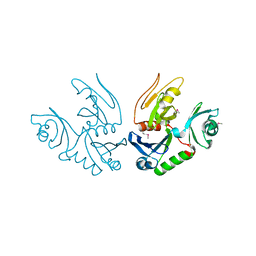 | | Cyto-Epsl: The Cytoplasmic Domain Of Epsl, An Inner Membrane Component Of The Type II Secretion System Of Vibrio Cholerae | | Descriptor: | General secretion pathway protein L | | Authors: | Abendroth, J, Murphy, P, Mushtaq, A, Sandkvist, M, Bagdasarian, M, Hol, W.G. | | Deposit date: | 2004-12-30 | | Release date: | 2005-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The X-ray Structure of the Type II Secretion System Complex Formed by the N-terminal Domain of EpsE and the Cytoplasmic Domain of EpsL of Vibrio cholerae.
J.Mol.Biol., 348, 2005
|
|
3NHJ
 
 | |
6KTJ
 
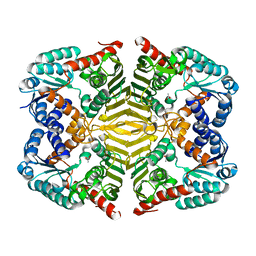 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, apo-form, from Paracoccus laeviglucosivorans | | Descriptor: | ACETATE ION, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
3NHU
 
 | |
1FU1
 
 | | CRYSTAL STRUCTURE OF HUMAN XRCC4 | | Descriptor: | ACETIC ACID, DNA REPAIR PROTEIN XRCC4 | | Authors: | Junop, M, Modesti, M, Guarne, A, Gellert, M, Yang, W. | | Deposit date: | 2000-09-13 | | Release date: | 2000-12-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Xrcc4 DNA repair protein and implications for end joining.
EMBO J., 19, 2000
|
|
1FY4
 
 | | FUSARIUM OXYSPORUM TRYPSIN AT ATOMIC RESOLUTION | | Descriptor: | GLY-ALA-ARG, GLYCEROL, SULFATE ION, ... | | Authors: | Rypniewski, W.R, Oestergaard, P, Noerregaard-Madsen, M, Dauter, M, Wilson, K.S. | | Deposit date: | 2000-09-28 | | Release date: | 2001-02-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Fusarium oxysporum trypsin at atomic resolution at 100 and 283 K: a study of ligand binding.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1YRQ
 
 | | Structure of the ready oxidized form of [NiFe]-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Cavazza, C, Matho, M, Faber, B.W, Roseboom, W, Albracht, S.P, Garcin, E, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2005-02-04 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases.
J.Biol.Inorg.Chem., 10, 2005
|
|
3O24
 
 | | Crystal structure of the brevianamide F prenyltransferase FtmPT1 from Aspergillus fumigatus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Brevianamide F prenyltransferase, CHLORIDE ION, ... | | Authors: | Jost, M, Zocher, G.E, Stehle, T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity.
J.Am.Chem.Soc., 132, 2010
|
|
2W01
 
 | |
1J0E
 
 | | ACC deaminase mutant reacton intermediate | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
2VYY
 
 | | Mutant Ala55Trp of Cerebratuls lacteus mini-hemoglobin | | Descriptor: | GLYCEROL, NEURAL HEMOGLOBIN, OXYGEN MOLECULE, ... | | Authors: | Salter, M.D, Nienhaus, K, Nienhaus, G.U, Dewilde, S, Moens, L, Pesce, A, Nardini, M, Bolognesi, M, Olson, J.S. | | Deposit date: | 2008-07-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Apolar Channel in Cerebratulus Lacteus Hemoglobin is the Route for O2 Entry and Exit.
J.Biol.Chem., 283, 2008
|
|
3NWE
 
 | | Rat COMT in complex with a methylated desoxyribose bisubstrate-containing inhibitor avoids hydroxyl group | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-(4-fluorophenyl)-2,3-dihydroxy-N-[(E)-3-[(2R,3R,4R,5R)-4-hydroxy-3-methyl-5-[6-(propylamino)purin-9-yl]oxolan-2-yl]prop-2-enyl]benzamide, CHLORIDE ION, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2010-07-09 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catechol-O-methyltransferase in complex with substituted 3'-deoxyribose bisubstrate inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2W7U
 
 | | SplA serine protease of Staphylococcus aureus (2.4A) | | Descriptor: | SERINE PROTEASE SPLA | | Authors: | Stec-Niemczyka, J, Pustelny, K, Kisielewska, M, Bista, M, Boulware, K.T, Stennicke, H.R, Thogersen, I.B, Daugherty, P.S, Enghild, J.J, Popowicz, G.M, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2008-12-30 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and Functional Characterization of Spla, an Exclusively Specific Protease of Staphylococcus Aureus.
Biochem.J., 419, 2009
|
|
2VSO
 
 | | Crystal Structure of a Translation Initiation Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-DEPENDENT RNA HELICASE EIF4A, EUKARYOTIC INITIATION FACTOR 4F SUBUNIT P150 | | Authors: | Schutz, P, Bumann, M, Oberholzer, A.E, Bieniossek, C, Altmann, M, Trachsel, H, Baumann, U. | | Deposit date: | 2008-04-28 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Yeast Eif4A-Eif4G Complex: An RNA-Helicase Controlled by Protein-Protein Interactions.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2A57
 
 | | Structure of 6,7-Dimthyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 6-carboxyethyl-7-oxo-8-ribityllumazine | | Descriptor: | 3-[8-((2S,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL)-2,4,7-TRIOXO-1,3,8-TRIHYDROPTERIDIN-6-YL]PROPANOIC ACID, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
2VWF
 
 | | Grb2 SH3C (2) | | Descriptor: | GRB2-ASSOCIATED-BINDING PROTEIN 2, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Harkiolaki, M, Tsirka, T, Feller, S.M. | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
2LZJ
 
 | | Refined solution structure and dynamics of First Catalytic Cysteine Half-domain from mouse E1 enzyme | | Descriptor: | Ubiquitin-like modifier-activating enzyme 1 | | Authors: | Jaremko, M, Jaremko, L, Nowakowski, M, Szczepanowski, R.H, Filipek, R, Wojciechowski, M, Bochtler, M, Ejchart, A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies of the first catalytic half-domain of ubiquitin activating enzyme.
J.Struct.Biol., 185, 2014
|
|
2VYQ
 
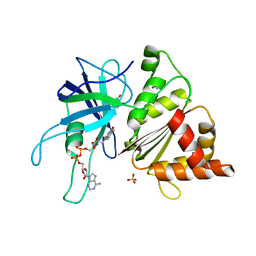 | | FERREDOXIN:NADP REDUCTASE MUTANT WITH THR 155 REPLACED BY GLY, ALA 160 REPLACED BY THR, LEU 263 REPLACED BY PRO AND TYR 303 REPLACED BY SER (T155G-A160T-L263P-Y303S) | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Peregrina, J.R, Medina, M. | | Deposit date: | 2008-07-28 | | Release date: | 2009-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein Motifs Involved in Coenzyme Interaction and Enzymatic Efficiency in Anabaena Ferredoxin-Nadp+ Reductase.
Biochemistry, 48, 2009
|
|
1HL3
 
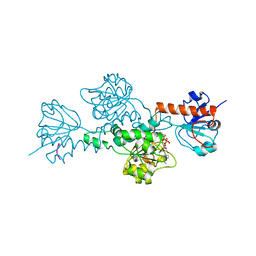 | | CtBP/BARS in ternary complex with NAD(H) and PIDLSKK peptide | | Descriptor: | C-TERMINAL BINDING PROTEIN 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PRO-ILE-ASP-LEU-SER-LYS-LYS PEPTIDE | | Authors: | Nardini, M, Spano, S, Cericola, C, Pesce, A, Massaro, A, Millo, E, Luini, A, Corda, D, Bolognesi, M. | | Deposit date: | 2003-03-13 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ctbp/Bars: A Dual-Function Protein Involved in Transcription Co-Repression and Golgi Membrane Fission
Embo J., 22, 2003
|
|
2CDM
 
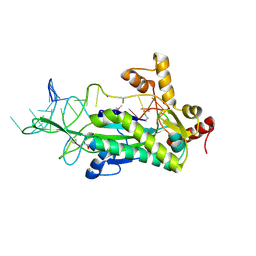 | | The structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site | | Descriptor: | 5'-D(*GP*CP*GP*CP*AP*CP*CP*GP*AP*AP *AP*GP*GP*TP*GP*CP*GP*TP*AP*TP*TP*GP*TP*CP*TP*AP*T)-3', SULFATE ION, TRWC | | Authors: | Boer, R, Russi, S, Guasch, A, Lucas, M, Blanco, A.G, Perez-Luque, R, Coll, M, de la Cruz, F. | | Deposit date: | 2006-01-25 | | Release date: | 2006-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unveiling the Molecular Mechanism of a Conjugative Relaxase: The Structure of Trwc Complexed with a 27-mer DNA Comprising the Recognition Hairpin and the Cleavage Site.
J.Mol.Biol., 358, 2006
|
|
1J2C
 
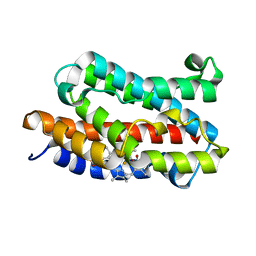 | | Crystal structure of rat heme oxygenase-1 in complex with biliverdin IXalpha-iron cluster | | Descriptor: | BILIVERDINE IX ALPHA, FE (III) ION, Heme Oxygenase-1 | | Authors: | Sugishima, M, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2002-12-29 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Rat Heme Oxygenase-1 in Complex with Biliverdin-Iron Chelate: CONFORMATIONAL CHANGE OF THE DISTAL HELIX DURING THE HEME CLEAVAGE REACTION.
J.Biol.Chem., 278, 2003
|
|
1JKA
 
 | | HUMAN LYSOZYME MUTANT WITH GLU 35 REPLACED BY ASP | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Muraki, M, Harata, K, Goda, S, Nagahora, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Importance of van der Waals contact between Glu 35 and Trp 109 to the catalytic action of human lysozyme.
Protein Sci., 6, 1997
|
|
1K5N
 
 | | HLA-B*2709 BOUND TO NONA-PEPTIDE M9 | | Descriptor: | GLYCEROL, beta-2-microglobulin, light chain, ... | | Authors: | Hulsmeyer, M, Hillig, R.C, Volz, A, Ruhl, M, Schroder, W, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2001-10-11 | | Release date: | 2002-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | HLA-B27 Subtypes Differentially Associated with Disease Exhibit Subtle Structural Alterations
J.Biol.Chem., 277, 2002
|
|
