6YHI
 
 | | Solution NMR Structure of APP G38L mutant TMD | | Descriptor: | Amyloid-beta precursor protein G38L mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
5RD5
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 27 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RDM
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 46 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RE3
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 12 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6YJB
 
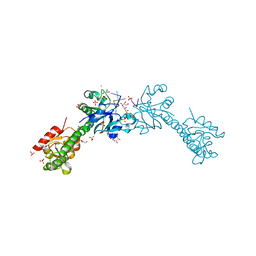 | | VcaM4I restriction endonuclease 5hmC-ssDNA complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*AP*(5HC)P*AP*G)-3'), GLYCEROL, ... | | Authors: | Pastor, M, Czapinska, H, Lutz, T, Helbrecht, I, Xu, S, Bochtler, M. | | Deposit date: | 2020-04-02 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the EVE-HNH endonuclease VcaM4I in the presence and absence of DNA.
Nucleic Acids Res., 49, 2021
|
|
2G4H
 
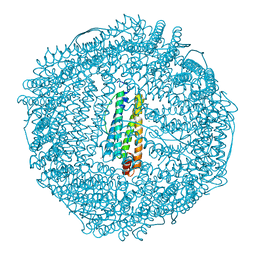 | | Anomalous substructure of apoferritin | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2G4L
 
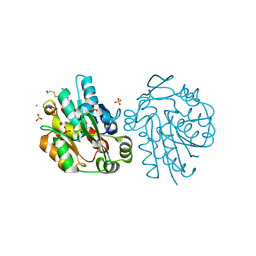 | | Anomalous substructure of hydroxynitrile lyase | | Descriptor: | (S)-acetone-cyanohydrin lyase, CHLORIDE ION, SULFATE ION | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2G4N
 
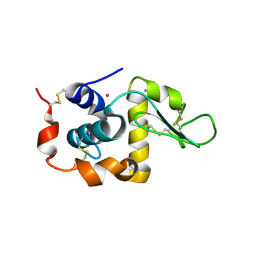 | | Anomalous substructure of alpha-lactalbumin | | Descriptor: | Alpha-lactalbumin, CALCIUM ION, POTASSIUM ION | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2G4Y
 
 | | structure of thaumatin at 2.0 A wavelength | | Descriptor: | D(-)-TARTARIC ACID, Thaumatin-1 | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5LBI
 
 | | Apo-structure of humanised RadA-mutant humRadA3 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5L6I
 
 | | Uba1 in complex with Ub-MLN4924 covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
8COY
 
 | | Structure of the catalytic domain of P. vivax Sub1 (triclinic crystal form) in complex with inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, SULFATE ION, ... | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5R0Z
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 13, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R1F
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 30, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6MQ2
 
 | |
8CP0
 
 | | Structure of the catalytic domain of P. vivax Sub1 (trigonal crystal form) | | Descriptor: | CALCIUM ION, subtilisin | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5R1W
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry C11, DMSO-free | | Descriptor: | Endothiapepsin, N-ethyl-N-(thiophene-2-carbonyl)-beta-alanine | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.077 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2D
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry H11, DMSO-free | | Descriptor: | 1-cyclopentyl-3-[[(2~{S})-oxolan-2-yl]methyl]urea, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.919 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2T
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 17, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.009 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
4O5Q
 
 | | Crystal Structure of the Alkylhydroperoxide Reductase AhpF from Escherichia coli | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkyl hydroperoxide reductase subunit F, CADMIUM ION, ... | | Authors: | Dip, P.V, Kamariah, N, Manimekalai, M.S.S, Balakrishna, A.M, Gruber, G. | | Deposit date: | 2013-12-20 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, mechanism and ensemble formation of the alkylhydroperoxide reductase subunits AhpC and AhpF from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8COZ
 
 | | Structure of the catalytic domain of P. vivax Sub1 (triclinic crystal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, SULFATE ION, ... | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.438 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5R3B
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 35, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3P
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 49, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.077 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RC3
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library F03a | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RCM
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 07 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
