7PSY
 
 | | X-ray crystal structure of perdeuterated LecB lectin in complex with perdeuterated fucose | | Descriptor: | CALCIUM ION, Fucose-binding lectin, SULFATE ION, ... | | Authors: | Gajdos, L, Blakeley, M.P, Haertlein, M, Forsyth, T.V, Devos, J.M, Imberty, A. | | Deposit date: | 2021-09-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Neutron crystallography reveals mechanisms used by Pseudomonas aeruginosa for host-cell binding.
Nat Commun, 13, 2022
|
|
8WNW
 
 | | the structure of PsaQ | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, PsaQ | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
7PX9
 
 | | Substrate-engaged mycobacterial Proteasome-associated ATPase - focused 3D refinement (state A) | | Descriptor: | AAA ATPase forming ring-shaped complexes, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jomaa, A, Kavalchuk, M, Weber-Ban, E. | | Deposit date: | 2021-10-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex.
Nat Commun, 13, 2022
|
|
8X8S
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with c-Myc fragment | | Descriptor: | Polyhedrin,Myc proto-oncogene protein | | Authors: | Kojima, M, Ueno, T, Abe, S, Hirata, K. | | Deposit date: | 2023-11-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | High-throughput structure determination of an intrinsically disordered protein using cell-free protein crystallization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7PXA
 
 | |
1EGE
 
 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
8WLG
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with c-Myc fragment | | Descriptor: | Polyhedrin,Myc proto-oncogene protein | | Authors: | Kojima, M, Ueno, T, Abe, S, Hirata, K. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | High-throughput structure determination of an intrinsically disordered protein using cell-free protein crystallization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X8V
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with c-Myc fragment | | Descriptor: | Polyhedrin,Myc proto-oncogene protein | | Authors: | Kojima, M, Ueno, T, Abe, S, Hirata, K. | | Deposit date: | 2023-11-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-throughput structure determination of an intrinsically disordered protein using cell-free protein crystallization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6JRE
 
 | | Structure of N-terminal domain of Plasmodium vivax p43 (PfNTD) solved by Co-SAD phasing | | Descriptor: | Aminoacyl-tRNA synthetase-interacting multifunctional protein p43, COBALT (II) ION | | Authors: | Manickam, Y, Harlos, K, Sharma, M, Gupta, S, Sharma, A. | | Deposit date: | 2019-04-03 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of the two domains that constitute the Plasmodium vivax p43 protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8WUV
 
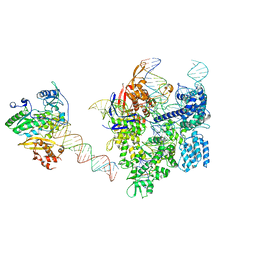 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 16-nt) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (50-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUU
 
 | | SpCas9-pegRNA-target DNA complex (pre-initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (34-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUS
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (termination) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (40-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
7PXC
 
 | | Substrate-engaged mycobacterial Proteasome-associated ATPase in complex with open-gate 20S CP - composite map (state A) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jomaa, A, Kavalchuk, M, Weber-Ban, E. | | Deposit date: | 2021-10-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex.
Nat Commun, 13, 2022
|
|
7PXB
 
 | | Substrate-engaged mycobacterial Proteasome-associated ATPase - focused 3D refinement (state B) | | Descriptor: | AAA ATPase forming ring-shaped complexes, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jomaa, A, Kavalchuk, M, Weber-Ban, E. | | Deposit date: | 2021-10-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex.
Nat Commun, 13, 2022
|
|
7PXD
 
 | | Substrate-engaged mycobacterial Proteasome-associated ATPase in complex with open-gate 20S CP - composite map (state B) | | Descriptor: | AAA ATPase forming ring-shaped complexes, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jomaa, A, Kavalchuk, M, Weber-Ban, E. | | Deposit date: | 2021-10-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex.
Nat Commun, 13, 2022
|
|
7UEG
 
 | |
7PT2
 
 | |
1E8L
 
 | | NMR solution structure of hen lysozyme | | Descriptor: | LYSOZYME | | Authors: | Schwalbe, H, Grimshaw, S.B, Spencer, A, Buck, M, Boyd, J, Dobson, C.M, Redfield, C, Smith, L.J. | | Deposit date: | 2000-09-27 | | Release date: | 2000-10-09 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | A refined solution structure of hen lysozyme determined using residual dipolar coupling data.
Protein Sci., 10, 2001
|
|
7PT1
 
 | |
7PT3
 
 | |
7PT4
 
 | | Actinobacterial 2-hydroxyacyl-CoA lyase (AcHACL) structure in complex with a covalently bound reaction intermediate as well as products formyl-CoA and acetone | | Descriptor: | 2-hydroxyacyl-CoA lyase, 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1R,11R,15S,17R)-19-[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]-1,11,15,17-TETRAHYDROXY-12,12-DIMETHYL-15,17-DIOXIDO-6,10-DIOXO-14,16,18-TRIOXA-2-THIA-5,9-DIAZA-15,17-DIPHOSPHANONADEC-1-YL}-5-(2-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, ACETONE, ... | | Authors: | Zahn, M, Rohwerder, T. | | Deposit date: | 2021-09-25 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanistic details of the actinobacterial lyase-catalyzed degradation reaction of 2-hydroxyisobutyryl-CoA.
J.Biol.Chem., 298, 2022
|
|
1EXG
 
 | | SOLUTION STRUCTURE OF A CELLULOSE BINDING DOMAIN FROM CELLULOMONAS FIMI BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | EXO-1,4-BETA-D-GLYCANASE | | Authors: | Xu, G.-Y, Ong, E, Gilkes, N.R, Kilburn, D.G, Muhandiram, D.R, Harris-Brandts, M, Carver, J.P, Kay, L.E, Harvey, T.S. | | Deposit date: | 1995-03-14 | | Release date: | 1995-06-03 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
8WT8
 
 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction intermediate) | | Descriptor: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
8WT7
 
 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange locked state | | Descriptor: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
1EH1
 
 | | RIBOSOME RECYCLING FACTOR FROM THERMUS THERMOPHILUS | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Toyoda, T, Tin, O.F, Ito, K, Fujiwara, T, Kumasaka, T, Yamamoto, M, Garber, M.B, Nakamura, Y. | | Deposit date: | 2000-02-18 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure combined with genetic analysis of the Thermus thermophilus ribosome recycling factor shows that a flexible hinge may act as a functional switch.
RNA, 6, 2000
|
|
