1K1Z
 
 | | Solution structure of N-terminal SH3 domain mutant(P33G) of murine Vav | | Descriptor: | vav | | Authors: | Ogura, K, Nagata, K, Horiuchi, M, Ebisui, E, Hasuda, T, Yuzawa, S, Nishida, M, Hatanaka, H, Inagaki, F. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-terminal SH3 domain of Vav and the recognition site for Grb2 C-terminal SH3 domain
J.BIOMOL.NMR, 22, 2002
|
|
2B8X
 
 | | Crystal structure of the interleukin-4 variant F82D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
1JKB
 
 | | HUMAN LYSOZYME MUTANT WITH GLU 35 REPLACED BY ALA | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Muraki, M, Harata, K, Goda, S, Nagahora, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Importance of van der Waals contact between Glu 35 and Trp 109 to the catalytic action of human lysozyme.
Protein Sci., 6, 1997
|
|
2B63
 
 | | Complete RNA Polymerase II-RNA inhibitor complex | | Descriptor: | 31-MER, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | Authors: | Kettenberger, H, Eisenfuehr, A, Brueckner, F, Theis, M, Famulok, M, Cramer, P. | | Deposit date: | 2005-09-30 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs
Nat.Struct.Mol.Biol., 13, 2006
|
|
2B8Z
 
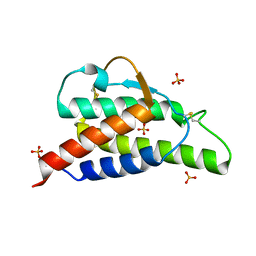 | | Crystal structure of the interleukin-4 variant R85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
1BJN
 
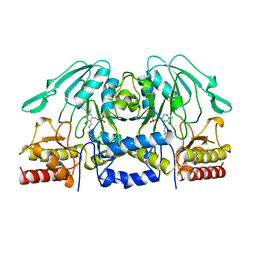 | |
1JNN
 
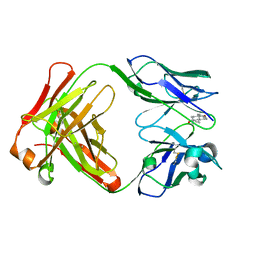 | | Crystal Structure of Fab-Estradiol Complexes | | Descriptor: | ESTRADIOL, MONOCLONAL ANTI-ESTRADIOL 17E12E5 IMMUNOGLOBULIN GAMMA-1 CHAIN, MONOCLONAL ANTI-ESTRADIOL 17E12E5 IMMUNOGLOBULIN KAPPA CHAIN | | Authors: | Monnet, C, Bettsworth, F, Stura, E.A, Le Du, M.-H, Menez, R, Derrien, L, Zinn-Justin, S, Gilquin, B, Sibai, G, Battail-Poirot, N, Jolivet, M, Menez, A, Arnaud, M, Ducancel, F, Charbonnier, J.B. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Highly specific anti-estradiol antibodies: structural characterisation and binding diversity.
J.Mol.Biol., 315, 2002
|
|
1RVW
 
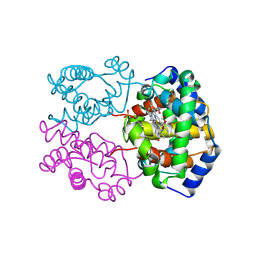 | | R STATE HUMAN HEMOGLOBIN [ALPHA V96W], CARBONMONOXY | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN, PHOSPHATE ION, ... | | Authors: | Puius, Y.A, Zou, M, Ho, N.T, Ho, C, Almo, S.C. | | Deposit date: | 1997-03-20 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel water-mediated hydrogen bonds as the structural basis for the low oxygen affinity of the blood substitute candidate rHb(alpha 96Val-->Trp).
Biochemistry, 37, 1998
|
|
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
1JU8
 
 | | Solution structure of Leginsulin, a plant hormon | | Descriptor: | Leginsulin | | Authors: | Yamazaki, T, Takaoka, M, Katoh, E, Hanada, K, Sakita, M, Sakata, K, Nishiuchi, Y, Hirano, H. | | Deposit date: | 2001-08-23 | | Release date: | 2003-06-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A possible physiological function and the tertiary structure of a 4-kDa peptide in legumes
EUR.J.BIOCHEM., 270, 2003
|
|
1D88
 
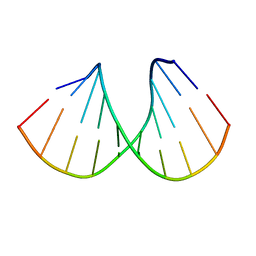 | |
1JQQ
 
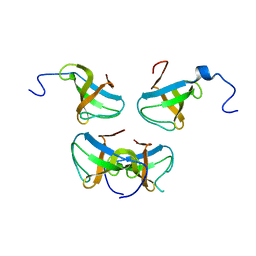 | | Crystal structure of Pex13p(301-386) SH3 domain | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PAS20 | | Authors: | Douangamath, A, Mayans, O, Barnett, P, Distel, B, Wilmanns, M. | | Deposit date: | 2001-08-08 | | Release date: | 2002-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Topography for Independent Binding of alpha-Helical and PPII-Helical Ligands to a Peroxisomal SH3 Domain
Mol.Cell, 10, 2002
|
|
2MK0
 
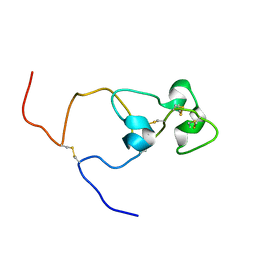 | | Structure of the PSCD4-domain of the cell wall protein pleuralin-1 from the diatom Cylindrotheca fusiformis | | Descriptor: | HEP200 protein | | Authors: | De Sanctis, S, Wenzler, M, Kroeger, N, Malloni, W.M, Sumper, M, Deutzmann, R, Zadravec, P, Brunner, E, Kremer, W, Kalbitzer, S.H.R. | | Deposit date: | 2014-01-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | PSCD Domains of Pleuralin-1 from the Diatom Cylindrotheca fusiformis: NMR Structures and Interactions with Other Biosilica-Associated Proteins.
Structure, 24, 2016
|
|
1RCY
 
 | | RUSTICYANIN (RC) FROM THIOBACILLUS FERROOXIDANS | | Descriptor: | COPPER (II) ION, RUSTICYANIN | | Authors: | Walter, R.L, Friedman, A.M, Ealick, S.E, Blake II, R.C, Proctor, P, Shoham, M. | | Deposit date: | 1996-04-10 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple wavelength anomalous diffraction (MAD) crystal structure of rusticyanin: a highly oxidizing cupredoxin with extreme acid stability.
J.Mol.Biol., 263, 1996
|
|
1JNH
 
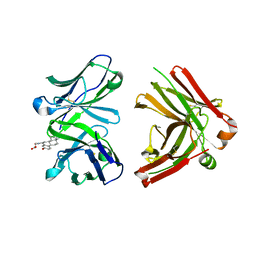 | | Crystal Structure of Fab-Estradiol Complexes | | Descriptor: | ESTRADIOL-6 CARBOXYL-METHYL-OXIME, monoclonal anti-estradiol 10G6D6 Fab heavy chain, monoclonal anti-estradiol 10G6D6 Fab light chain | | Authors: | Monnet, C, Bettsworth, F, Stura, E.A, Le Du, M.-H, Menez, R, Derrien, L, Zinn-Justin, S, Gilquin, B, Sibai, G, Battail-Poirot, N, Jolivet, M, Menez, A, Arnaud, M, Ducancel, F, Charbonnier, J.B. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Highly specific anti-estradiol antibodies: structural characterisation and binding diversity.
J.Mol.Biol., 315, 2002
|
|
2C98
 
 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
2C99
 
 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
2CCJ
 
 | | Crystal structure of S. aureus thymidylate kinase complexed with thymidine monophosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
2C96
 
 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
1K37
 
 | | NMR Structure of human Epiregulin | | Descriptor: | Epiregulin | | Authors: | Sato, K, Miura, K, Tada, M, Aizawa, T, Miyamoto, K, Kawano, K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of epiregulin and the effect of its C-terminal domain for receptor binding affinity
Febs Lett., 553, 2003
|
|
1JKK
 
 | | 2.4A X-RAY STRUCTURE OF TERNARY COMPLEX OF A CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE WITH ATP ANALOGUE AND MG. | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-07-12 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
1JWG
 
 | | VHS Domain of human GGA1 complexed with cation-independent M6PR C-terminal Peptide | | Descriptor: | ADP-ribosylation factor binding protein GGA1, Cation-independent mannose-6-phosphate receptor, IODIDE ION | | Authors: | Shiba, T, Takatsu, H, Nogi, T, Matsugaki, N, Kawasaki, M, Igarashi, N, Suzuki, M, Kato, R, Earnest, T, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for recognition of acidic-cluster dileucine sequence by GGA1.
Nature, 415, 2002
|
|
2CKS
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THERMOBIFIDA FUSCA ENDOGLUCANASE CEL5A (E5) | | Descriptor: | BENZAMIDINE, ENDOGLUCANASE E-5, SODIUM ION, ... | | Authors: | Berglund, G.I, Gualfetti, P.J, Requadt, C, Gross, L.S, Bergfors, T, Shaw, A, Saldajeno, M, Mitchinson, C, Sandgren, M. | | Deposit date: | 2006-04-21 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Catalytic Domain of Thermobifida Fusca Endoglucanase Cel5A in Complex with Cellotetraose
To be Published
|
|
1K23
 
 | | Inorganic Pyrophosphatase (Family II) from Bacillus subtilis | | Descriptor: | MANGANESE (II) ION, Manganese-dependent inorganic pyrophosphatase | | Authors: | Ahn, S, Milner, A.J, Futterer, K, Konopka, M, Ilias, M, Young, T.W, White, S.A. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The "open" and "closed" structures of the type-C inorganic pyrophosphatases from Bacillus subtilis and Streptococcus gordonii.
J.Mol.Biol., 313, 2001
|
|
1K36
 
 | | NMR Structure of human Epiregulin | | Descriptor: | Epiregulin | | Authors: | Sato, K, Miura, K, Tada, M, Aizawa, T, Miyamoto, K, Kawano, K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-09-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of epiregulin and the effect of its C-terminal domain for receptor binding affinity
Febs Lett., 553, 2003
|
|
