8XEQ
 
 | |
8HTY
 
 | | Candida boidinii Formate Dehydrogenase Crystal Structure at 1.4 Angstrom Resolution | | Descriptor: | Formate dehydrogenase, SULFATE ION | | Authors: | Gul, M, Yuksel, B, Bulut, H, DeMirci, H. | | Deposit date: | 2022-12-22 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural analysis of wild-type and Val120Thr mutant Candida boidinii formate dehydrogenase by X-ray crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3QIA
 
 | | Crystal structure of P-loop G237A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-26 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
8XGW
 
 | |
3QQQ
 
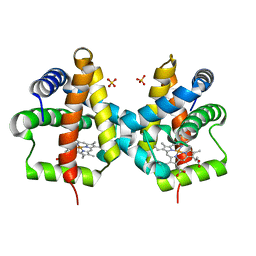 | | Crystal structure of non-symbiotic plant hemoglobin from Trema tomentosa | | Descriptor: | Non-symbiotic hemoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kakar, S, Sturms, R, Savage, A, Nix, J.C, Dispirito, A, Hargrove, M.S. | | Deposit date: | 2011-02-16 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structures of Parasponia and Trema hemoglobins: differential heme coordination is linked to quaternary structure.
Biochemistry, 50, 2011
|
|
6AA4
 
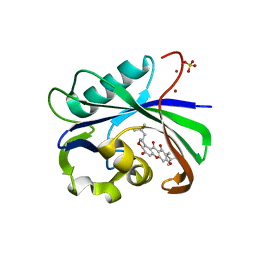 | | Crystal structure of MTH1 in complex with alpha-mangostin (cocktail No. 9) | | Descriptor: | 1,3,6-trihydroxy-7-methoxy-2,8-bis(3-methylbut-2-en-1-yl)-9H-xanthen-9-one, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION, ... | | Authors: | Yokoyama, T, Kitakami, R, Mizuguchi, M. | | Deposit date: | 2018-07-17 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a new class of MTH1 inhibitor by X-ray crystallographic screening.
Eur J Med Chem, 167, 2019
|
|
5ZWP
 
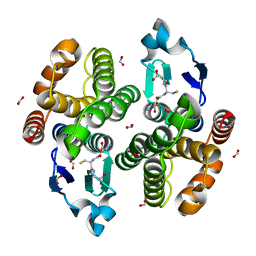 | |
8SX0
 
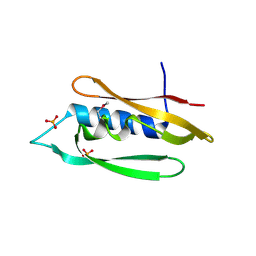 | | Bordetella filamentous hemagglutinin (FhaB) C-terminal domain | | Descriptor: | CALCIUM ION, Filamentous hemagglutinin, GLYCEROL, ... | | Authors: | Cuthbert, B.J, Goulding, C.W, Hayes, C.S, Costello, M.S. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bordetella filamentous hemagglutinin (FhaB) C-terminal domain
To Be Published
|
|
8I2J
 
 | |
8SW6
 
 | | Protein Phosphatase 1 in complex with PP1-specific Phosphatase targeting peptide (PhosTAP) version 3 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PP1-specific Phosphatase-Targeting Peptide version 3, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A protein phosphatase 1 specific phos phatase ta rgeting p eptide (PhosTAP) to identify the PP1 phosphatome.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8I1Y
 
 | | The structure of E. coli TrpRS bound with a chemical fragment | | Descriptor: | 5-ethanoylthiophene-2-carbonitrile, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
8I27
 
 | |
8I2A
 
 | |
8I4I
 
 | |
7XM1
 
 | | Cryo-EM structure of mTIP60-Ba (metal-ion induced TIP60 (K67E) complex with barium ions | | Descriptor: | BARIUM ION, TIP60 K67E mutant | | Authors: | Ohara, N, Kawakami, N, Arai, R, Adachi, N, Moriya, T, Kawasaki, M, Miyamoto, K. | | Deposit date: | 2022-04-24 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J.Am.Chem.Soc., 145, 2023
|
|
8I1W
 
 | |
8I1Z
 
 | | E. coli tryptophanyl-tRNA synthetase bound with a chemical fragment | | Descriptor: | 1-(2,3-dihydro-1-benzofuran-5-yl)ethanone, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
8I2M
 
 | |
8I2C
 
 | |
8SW5
 
 | | Protein Phosphatase 1 in complex with PP1-specific Phosphatase targeting peptide (PhosTAP) version 1 | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PP1-specific Phosphatase-Targeting Peptide version 1, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A protein phosphatase 1 specific phos phatase ta rgeting p eptide (PhosTAP) to identify the PP1 phosphatome.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8I2L
 
 | |
8IDS
 
 | | Crystal structure of Bacillus sp. AHU2216 GH13_31 Alpha-glucosidase E256Q/N258P in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Auiewiriyanukul, W, Saburi, W, Yu, J, Kato, K, Yao, M, Mori, H. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 alpha-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on beta → alpha Loop 5.
Molecules, 28, 2023
|
|
8IDQ
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus with xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
8IGR
 
 | | Cryo-EM structure of CII-dependent transcription activation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhao, M, Gao, B, Wen, A, Feng, Y, Lu, Y. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of lambda CII-dependent transcription activation.
Structure, 31, 2023
|
|
8IGS
 
 | | Cryo-EM structure of RNAP-promoter open complex at lambda promoter PRE | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhao, M, Gao, B, Wen, A, Feng, Y, Lu, Y. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of lambda CII-dependent transcription activation.
Structure, 31, 2023
|
|
