6YYK
 
 | | Crystal Structure of 1,5-dimethylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 1,5-dimethyl-3~{H}-indol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
1W8D
 
 | | Binary structure of human DECR. | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, MITOCHONDRIAL PRECURSOR, GLYCEROL, ... | | Authors: | Alphey, M.S, Byres, E, Hunter, W.N. | | Deposit date: | 2004-09-20 | | Release date: | 2004-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Reactivity of Human Mitochondrial 2,4-Dienoyl-Coa Reductase: Enzyme-Ligand Interactions in a Distinctive Short-Chain Reductase Active Site
J.Biol.Chem., 280, 2005
|
|
6YYF
 
 | | Crystal Structure of 5-chloroindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 5-chloranyl-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6YYG
 
 | | Crystal Structure of 5-(trifluoromethoxy)indoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 5-(trifluoromethyloxy)-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
2V34
 
 | | IspE in complex with cytidine and ligand | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, CHLORIDE ION, ... | | Authors: | Sgraja, T, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-06-11 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Aquifex Aeolicus 4-Diphosphocytidyl-2C-Methyl-D-Erythritol Kinase -Ligand Recognition in a Template for Antimicrobial Drug Discovery.
FEBS J., 275, 2008
|
|
7OLF
 
 | |
6Z0E
 
 | |
6Z0Y
 
 | |
6Z0Z
 
 | | Human wtSTING in complex with 3',3'-c-(2'FdAMP-2'FdAMP) | | Descriptor: | 2'-fluoro-,3',3'-c-di-AMP, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6Z0X
 
 | |
2V2Z
 
 | | IspE in complex with ADP and CDPME | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL, 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sgraja, T, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-06-08 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of Aquifex Aeolicus 4-Diphosphocytidyl-2C-Methyl-D-Erythritol Kinase -Ligand Recognition in a Template for Antimicrobial Drug Discovery.
FEBS J., 275, 2008
|
|
2V8P
 
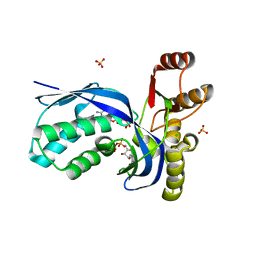 | | IspE in complex with ADP and CDP | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL KINASE, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Sgraja, T, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-08-13 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of Aquifex Aeolicus 4-Diphosphocytidyl-2C-Methyl-D-Erythritol Kinase -Ligand Recognition in a Template for Antimicrobial Drug Discovery.
FEBS J., 275, 2008
|
|
1W0D
 
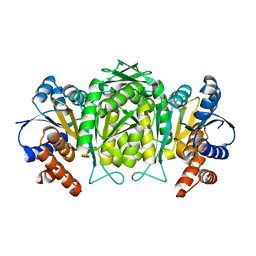 | | The high resolution structure of Mycobacterium tuberculosis LeuB (Rv2995c) | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | Authors: | Singh, R.K, Kefala, G, Janowski, R, Mueller-Dieckmann, C, Weiss, M.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-06-03 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The High Resolution Structure of Leub (Rv2995C) from Mycobacterium Tuberculosis
J.Mol.Biol., 346, 2005
|
|
5ZA8
 
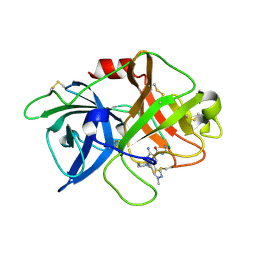 | | uPA-BB2-27F | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(1-methylpyrazol-4-yl)pyrazine-2-carboxamide, Urokinase-type plasminogen activator chain B | | Authors: | Buckley, B.J, Jiang, L.G, Huang, M.D, Kelso, M.J, Ranson, M. | | Deposit date: | 2018-02-06 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 6-Substituted Hexamethylene Amiloride (HMA) Derivatives as Potent and Selective Inhibitors of the Human Urokinase Plasminogen Activator for Use in Cancer.
J. Med. Chem., 61, 2018
|
|
5ZAF
 
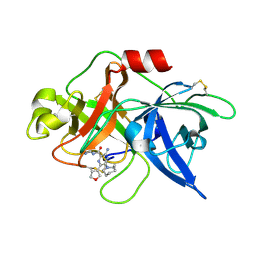 | | uPA-BB2-28F | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(furan-3-yl)pyrazine-2-carboxamide, Urokinase-type plasminogen activator chain B | | Authors: | Buckley, B.J, Jiang, L.G, Huang, M.D, Kelso, M.J, Ranson, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 6-Substituted Hexamethylene Amiloride (HMA) Derivatives as Potent and Selective Inhibitors of the Human Urokinase Plasminogen Activator for Use in Cancer.
J. Med. Chem., 61, 2018
|
|
7OVP
 
 | |
6XTC
 
 | |
5NW1
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(cyclobutanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 18) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-(cyclobutylcarbonylamino)-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
7OOZ
 
 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 6-benzyloxo-2-chloropurine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-benzyloxo-2-chloropurine, GLYCEROL, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7OP9
 
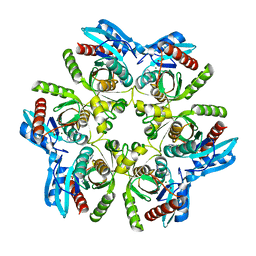 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 2,6-dichloropurine | | Descriptor: | 2,6-bis(chloranyl)-7H-purine, IMIDAZOLE, MAGNESIUM ION, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7OPA
 
 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 6-benzylthiopurine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-benzylthio-2-chloropurine, GLYCEROL, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
4PPH
 
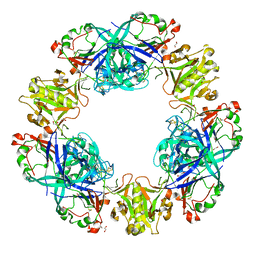 | | Crystal structure of conglutin gamma, a unique basic 7S globulin from lupine seeds | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Czubinski, J, Barciszewski, J, Gilski, M, Lampart-Szczapa, E, Jaskolski, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Structure of gamma-conglutin: insight into the quaternary structure of 7S basic globulins from legumes.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7OOY
 
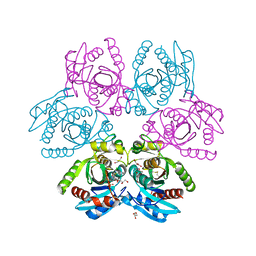 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 6-benzylthio-2-chloropurine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-benzylthio-2-chloropurine, GLYCEROL, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7OYJ
 
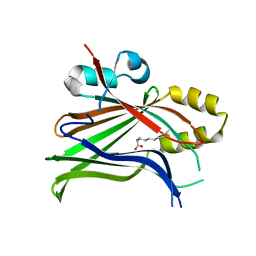 | |
1W73
 
 | | Binary structure of human DECR solved by SeMet SAD. | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Alphey, M.S, Byres, E, Hunter, W.N. | | Deposit date: | 2004-08-27 | | Release date: | 2004-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Reactivity of Human Mitochondrial 2,4-Dienoyl-Coa Reductase: Enzyme-Ligand Interactions in a Distinctive Short-Chain Reductase Active Site
J.Biol.Chem., 280, 2005
|
|
