7ZMT
 
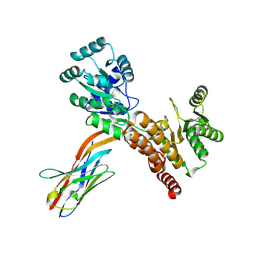 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G5-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
2OXC
 
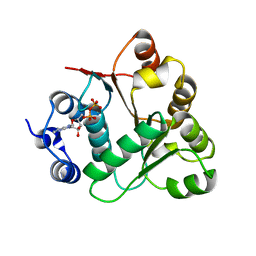 | | Human DEAD-box RNA helicase DDX20, DEAD domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable ATP-dependent RNA helicase DDX20 | | Authors: | Karlberg, T, Ogg, D, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Lehtio, L, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-20 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human DEAD-box RNA helicase DDX20
To be Published
|
|
7ZMP
 
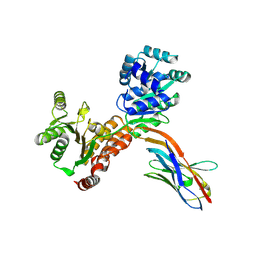 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-055 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-055, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.626 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMR
 
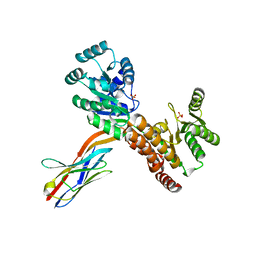 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G2*-011 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G2*-011, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
2KM6
 
 | | NMR structure of the NLRP7 Pyrin domain | | Descriptor: | NACHT, LRR and PYD domains-containing protein 7 | | Authors: | Pinheiro, A, Proell, M, Schwarzenbacher, R, Peti, W. | | Deposit date: | 2009-07-21 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the NLRP7 pyrin domain: insight into pyrin-pyrin-mediated effector domain signaling in innate immunity.
J.Biol.Chem., 285, 2010
|
|
2KYY
 
 | | Solution NMR Structure of the N-terminal Domain of Putative ATP-dependent DNA Helicase RecG-related Protein from Nitrosomonas europaea, Northeast Structural Genomics Consortium Target NeR70A | | Descriptor: | Possible ATP-dependent DNA helicase RecG-related protein | | Authors: | Eletsky, A, Mills, J.L, Lee, H, Maglaqui, M, Ciccosanti, C, Hamilton, K, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the N-terminal Domain of Putative ATP-dependent DNA Helicase RecG-related Protein from Nitrosomonas europaea
To be Published
|
|
3WXL
 
 | | Crystal structure of trypanosoma brucei gambiense glycerol kinase complex with adp, mg2+, and glycerol | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Glycerol kinase, ... | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Kido, Y, Tsuge, C, Nara, T, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Kita, K, Harada, S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the reverse reaction of African human trypanosomes glycerol kinase.
Mol.Microbiol., 94, 2014
|
|
2KA5
 
 | | NMR Structure of the protein TM1081 | | Descriptor: | Putative anti-sigma factor antagonist TM_1081 | | Authors: | Serrano, P, Geralt, M, Mohanty, B, Pedrini, B, Horst, R, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7ZMQ
 
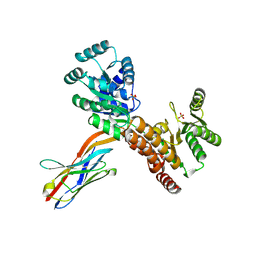 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G2*-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G2*-006, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
2KHV
 
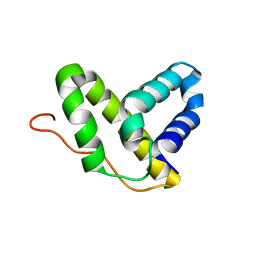 | | Solution NMR structure of protein Nmul_A0922 from Nitrosospira multiformis. Northeast Structural Genomics Consortium target NmR38B. | | Descriptor: | Phage integrase | | Authors: | Wu, Y, Mills, J.L, Wang, D, Ciccosanti, C, Sukumaran, D.K, Jiang, M, Garcia, E, Nair, R, Rost, B, Swapna, G.V.T, Acton, T.B, Xiao, R, Everett, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-13 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein Nmul_A0922 from Nitrosospira multiformis. Northeast Structural Genomics Consortium target NmR38B.
To be Published
|
|
7ZML
 
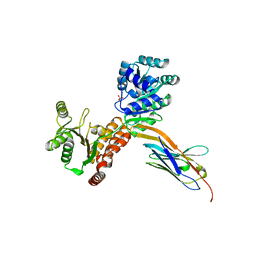 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G1-001 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G1-001, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
3WXJ
 
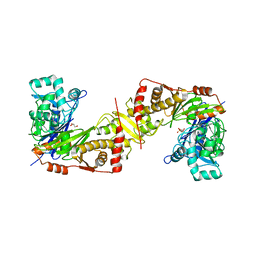 | | Crystal structure of trypanosoma brucei gambiense glycerol kinase in complex with glycerol 3-phosphate | | Descriptor: | GLYCEROL, Glycerol kinase, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Kido, Y, Tsuge, C, Nara, T, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Kita, K, Harada, S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for the reverse reaction of African human trypanosomes glycerol kinase.
Mol.Microbiol., 94, 2014
|
|
2KPA
 
 | | Specific motifs of the V-ATPase a2-subunit isoform interact with catalytic and regulatory domains of ARNO | | Descriptor: | ARNO(375-400) | | Authors: | Merkulova, M, Bakulina, A, Thaker, Y.R, Gruber, G, Marshansky, V. | | Deposit date: | 2009-10-11 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Specific motifs of the V-ATPase a2-subunit isoform interact with catalytic and regulatory domains of ARNO
Biochim.Biophys.Acta, 2010
|
|
2KIJ
 
 | | Solution structure of the Actuator domain of the copper-transporting ATPase ATP7A | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Nushi, F, Natile, G, Rosato, A. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the actuator domain of ATP7A and ATP7B, the Menkes and Wilson disease proteins
Biochemistry, 48, 2009
|
|
7ZMM
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G2-001 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G2-001, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMN
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-048 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-048, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
2KAT
 
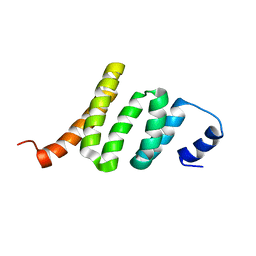 | | Solution structure of protein BPP2914 from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR206 | | Descriptor: | uncharacterized protein | | Authors: | Wu, Y, Mills, J.L, Wang, D, Jiang, M, Foote, E.L, Xiao, R, Sathyamoorthy, B, Sukumaran, D, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein BPP2914 from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR206
To be Published
|
|
4EHR
 
 | | Crystal structure of Bcl-Xl complex with 4-(5-butyl-3-(hydroxymethyl)-1-phenyl-1h-pyrazol-4-yl)-3-(3,4-dihydro-2(1h)-isoquinolinylcarbonyl)-n-((2-(trimethylsilyl)ethyl)sulfonyl)benzamide | | Descriptor: | 4-[5-butyl-3-(hydroxymethyl)-1-phenyl-1H-pyrazol-4-yl]-3-(3,4-dihydroisoquinolin-2(1H)-ylcarbonyl)-N-{[2-(trimethylsilyl)ethyl]sulfonyl}benzamide, Bcl-2-like protein 1, IMIDAZOLE | | Authors: | Schroeder, G.M, Wei, D, Banfi, P, Cai, Z, Lippy, J, Menichincheri, M, Modugno, M, Naglich, J, Penhallow, B, Perez, H.L, Sack, J, Schmidt, R.J, Tebben, A, Yan, C, Zhang, L, Galvani, A, Lombardo, L.J, Borzilleri, R.M. | | Deposit date: | 2012-04-03 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyrazole and pyrimidine phenylacylsulfonamides as dual Bcl-2/Bcl-xL antagonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2KIS
 
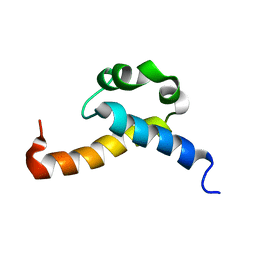 | | Solution structure of CA150 FF1 domain and FF1-FF2 interdomain linker | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Murphy, J.M, Hansen, D, Wiesner, S, Muhandiram, D, Borg, M, Smith, M.J, Sicheri, F, Kay, L.E, Forman-Kay, J.D, Pawson, T. | | Deposit date: | 2009-05-08 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural studies of FF domains of the transcription factor CA150 provide insights into the organization of FF domain tandem arrays.
J.Mol.Biol., 393, 2009
|
|
3W77
 
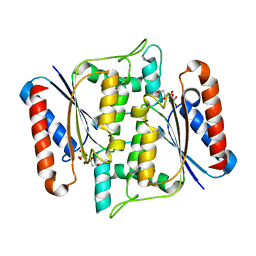 | | Crystal Structure of azoreductase AzrA | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Ogata, D, Yu, J, Ooi, T, Yao, M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WHC
 
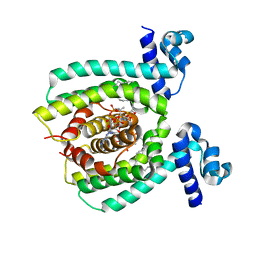 | |
2K97
 
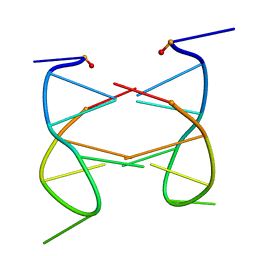 | | Dimeric solution structure of the cyclic octamer d(pCGCTCCGT) | | Descriptor: | 5'-D(P*CP*GP*CP*TP*CP*CP*GP*T)-3' | | Authors: | Viladoms, J, Escaja, N, Frieden, M, Gomez-Pinto, I, Pedroso, E, Gonzalez, C. | | Deposit date: | 2008-09-30 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Self-association of short DNA loops through minor groove C:G:G:C tetrads.
Nucleic Acids Res., 37, 2009
|
|
2K9Z
 
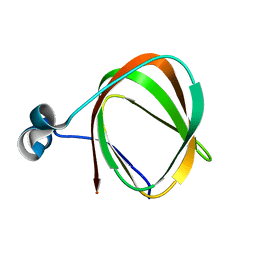 | | NMR structure of the protein TM1112 | | Descriptor: | uncharacterized protein TM1112 | | Authors: | Mohanty, B, Pedrini, B, Serrano, P, Geralt, M, Horst, R, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures for the proteins TM1112 and TM1367.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3WI6
 
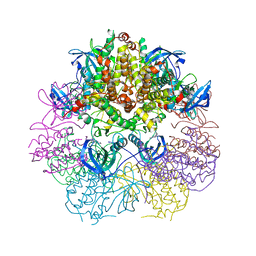 | | Crystal structure of MAPKAP Kinase-2 (MK2) in complex with non-selective inhibitor | | Descriptor: | MAP kinase-activated protein kinase 2, N-[(3S)-piperidin-3-yl]-7,8-dihydro-6H-pyrazolo[1,5-a]pyrrolo[3,2-e]pyrimidin-5-amine | | Authors: | Fujino, A, Fukushima, K, Kubota, T, Matsumoto, Y, Takimoto-Kamimura, M. | | Deposit date: | 2013-09-06 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of the beta-form of human MK2 in complex with the non-selective kinase inhibitor TEI-L03090
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3W8F
 
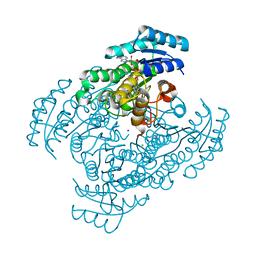 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and an inhibitor malonate | | Descriptor: | CHLORIDE ION, D-3-hydroxybutyrate dehydrogenase, MALONIC ACID, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray diffraction of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and malonate
To be Published
|
|
