6SE6
 
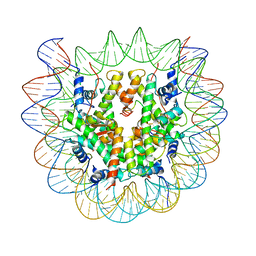 | | Class2 : CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | Centromere protein C, DNA (145-MER), Histone H2A type 2-A, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
4NF9
 
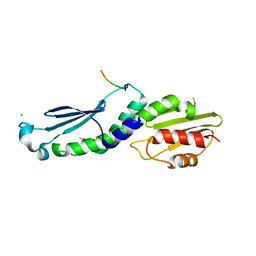 | | Structure of the Knl1/Nsl1 complex | | Descriptor: | CHLORIDE ION, Kinetochore-associated protein NSL1 homolog, Protein CASC5 | | Authors: | Petrovic, A, Mosalaganti, S, Keller, J, Mattiuzzo, M, Overlack, K, Wohlgemuth, S, Pasqualato, S, Raunser, S, Musacchio, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modular Assembly of RWD Domains on the Mis12 Complex Underlies Outer Kinetochore Organization.
Mol.Cell, 53, 2014
|
|
1JR3
 
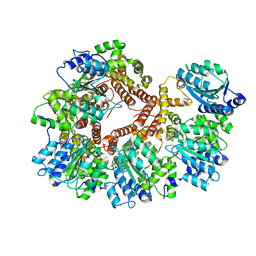 | | Crystal Structure of the Processivity Clamp Loader Gamma Complex of E. coli DNA Polymerase III | | Descriptor: | DNA polymerase III subunit gamma, DNA polymerase III, delta subunit, ... | | Authors: | Jeruzalmi, D, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2001-08-10 | | Release date: | 2001-09-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
6SKY
 
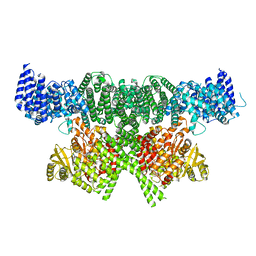 | | FAT and kinase domain of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
1JRW
 
 | | Solution Structure of dAATAA DNA Bulge | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*CP*GP*AP*AP*TP*AP*AP*GP*CP*TP*AP*CP*G)-3' | | Authors: | Gollmick, F.A, Lorenz, M, Dornberger, U, von Langen, J, Diekmann, S, Fritzsche, H. | | Deposit date: | 2001-08-15 | | Release date: | 2002-08-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dAATAA and dAAUAA DNA bulges.
Nucleic Acids Res., 30, 2002
|
|
6SF5
 
 | | Mn-containing form of the ribonucleotide reductase NrdB protein from Leeuwenhoekiella blandensis | | Descriptor: | MANGANESE (II) ION, Ribonucleoside-diphosphate reductase, beta subunit 1 | | Authors: | Hasan, M, Rozman Grinberg, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2019-08-01 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Class Id ribonucleotide reductase utilizes a Mn2(IV,III) cofactor and undergoes large conformational changes on metal loading.
J.Biol.Inorg.Chem., 24, 2019
|
|
3F7T
 
 | | Structure of active IspH shows a novel fold with a [3Fe-4S] cluster in the catalytic centre | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, PHOSPHATE ION, ... | | Authors: | Graewert, T, Eppinger, J, Rohdich, F, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2008-11-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of active IspH enzyme from Escherichia coli provides mechanistic insights into substrate reduction.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4NJB
 
 | | Crystal structure of the complex of lactoperoxidase from bovine with 3,3-oxydipyridine at 2.31 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Sirohi, H.V, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-11-09 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of the complex of lactoperoxidase from bovine with 3,3-oxydipyridine at 2.31 A resolution
To be Published
|
|
3QCQ
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-(3-Amino-1H-indazol-6-yl)-N4-ethyl-2,4-pyrimidinediamine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-(3-amino-2H-indazol-6-yl)-N~4~-ethylpyrimidine-2,4-diamine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
6SNI
 
 | | Cryo-EM structure of nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab | | Descriptor: | 6AG9-Fab heavy chain, 6AG9-Fab light chain, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Bloch, J.S, Pesciullesi, G, Boilevin, J, Nosol, K, Irobalieva, R.N, Darbre, T, Aebi, M, Kossiakoff, A.A, Reymond, J.L, Locher, K.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and mechanism of the ER-based glucosyltransferase ALG6.
Nature, 579, 2020
|
|
4NHZ
 
 | | Crystal structure of glutathione transferase BBTA-3750 from Bradyrhizobium sp., Target EFI-507290, with one glutathione bound | | Descriptor: | GLUTATHIONE, Putative glutathione S-transferase enzyme with thioredoxin-like domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal Structure of Glutathione Transferase Bbta-3750 from Bradyrhizobium Sp., Target Efi-507290
To be Published
|
|
6SB0
 
 | | cryo-EM structure of mTORC1 bound to PRAS40-fused active RagA/C GTPases | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Proline-rich AKT1 substrate 1, ... | | Authors: | Anandapadamanaban, M, Berndt, A, Masson, G.R, Perisic, O, Williams, R.L. | | Deposit date: | 2019-07-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science, 366, 2019
|
|
3FJ9
 
 | |
4NKH
 
 | | Crystal structure of SspH1 LRR domain | | Descriptor: | E3 ubiquitin-protein ligase sspH1 | | Authors: | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | Deposit date: | 2013-11-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
6F6K
 
 | |
1JN2
 
 | | Crystal Structure of meso-tetrasulphonatophenyl porphyrin complexed with Concanavalin A | | Descriptor: | 5,10,15,20-TETRAKIS(4-SULPFONATOPHENYL)-21H,23H-PORPHINE, CALCIUM ION, Concanavalin-A, ... | | Authors: | Goel, M, Jain, D, Kaur, K.J, Kenoth, R, Maiya, B.G, Swamy, M.J, Salunke, D.M. | | Deposit date: | 2001-07-22 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional equality in the absence of structural similarity: an added dimension to
molecular mimicry
J.Biol.Chem., 276, 2000
|
|
4J49
 
 | | PylD holoenzyme soaked with L-lysine-Ne-D-ornithine | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Quitterer, F, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2013-02-06 | | Release date: | 2013-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Reaction Mechanism of Pyrrolysine Synthase (PylD).
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3QE4
 
 | | An evolved aminoacyl-tRNA Synthetase with atypical polysubstrate specificity | | Descriptor: | 4-cyano-L-phenylalanine, Tyrosyl-tRNA synthetase | | Authors: | Young, D.D, Young, T.S, Jahnz, M, Ahmad, I, Spraggon, G, Schultz, P.G. | | Deposit date: | 2011-01-19 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Evolved Aminoacyl-tRNA Synthetase with Atypical Polysubstrate Specificity .
Biochemistry, 50, 2011
|
|
6SEE
 
 | | Class2A : CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | Centromere protein C, DNA (145-MER), Histone H2A type 2-A, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
3FKZ
 
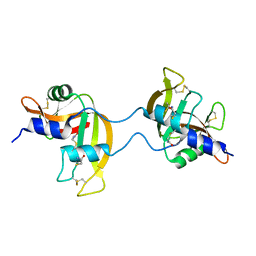 | | X-ray structure of the non covalent swapped form of the S16G/T17N/A19P/A20S/K31C/S32C mutant of bovine pancreatic ribonuclease | | Descriptor: | Ribonuclease pancreatic | | Authors: | Merlino, A, Russo Krauss, I, Perillo, M, Mattia, C.A, Ercole, C, Picone, D, Vergara, A, Sica, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Toward an antitumor form of bovine pancreatic ribonuclease: The crystal structure of three noncovalent dimeric mutants
Biopolymers, 91, 2009
|
|
1JIR
 
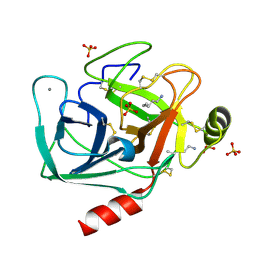 | | Crystal Structure of Trypsin Complex with Amylamine in Cyclohexane | | Descriptor: | AMYLAMINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Wu, G, Zhu, G, Huang, Q, Qian, M, Tang, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2001-07-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of beta-Trypsin Complex with Amylamine in Cyclohexane
To be Published
|
|
6HX5
 
 | |
6SF4
 
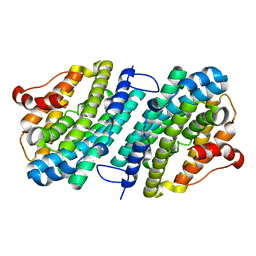 | | Apo form of the ribonucleotide reductase NrdB protein from Leeuwenhoekiella blandensis | | Descriptor: | Ribonucleoside-diphosphate reductase, beta subunit 1 | | Authors: | Hasan, M, Rozman Grinberg, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Class Id ribonucleotide reductase utilizes a Mn2(IV,III) cofactor and undergoes large conformational changes on metal loading.
J.Biol.Inorg.Chem., 24, 2019
|
|
2M86
 
 | | Solution structure of Hdm2 with engineered cyclotide | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, MCo-PMI | | Authors: | Majumder, S, Ji, Y, Millard, M, Borra, R, Bi, T, Elnagar, A.Y, Neamati, N, Camarero, J.A. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | In Vivo Activation of the p53 Tumor Suppressor Pathway by an Engineered Cyclotide.
J.Am.Chem.Soc., 135, 2013
|
|
4J99
 
 | |
