6SLD
 
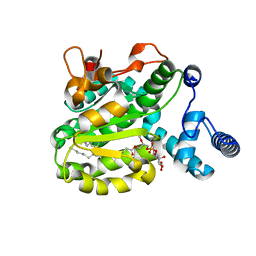 | | Structure of the Phosphatidylcholine Binding Mutant of Yeast Sec14 Homolog Sfh1 (S175I,T177I) in Complex with Phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CRAL-TRIO domain-containing protein YKL091C | | Authors: | Berger, J, Fitz, M, Johnen, P, Shanmugaratnam, S, Hocker, B, Stiel, A.C, Schaaf, G. | | Deposit date: | 2019-08-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Phosphatidylcholine Binding Mutant of Yeast Sec14 Homolog Sfh1 (S175I,T177I) in Complex with Phosphatidylinositol
To Be Published
|
|
5O2Y
 
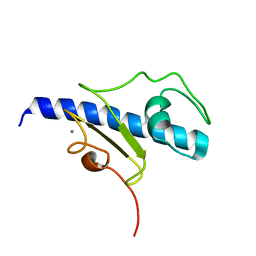 | | NMR structure of the calcium bound form of PulG, major pseudopilin from Klebsiella oxytoca T2SS | | Descriptor: | CALCIUM ION, General secretion pathway protein G | | Authors: | Lopez-Castilla, A, Bardiaux, B, Vitorge, B, Thomassin, J.-L, Zheng, W, Yu, X, Egelman, E.H, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2017-05-23 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the calcium-dependent type 2 secretion pseudopilus.
Nat Microbiol, 2, 2017
|
|
7LF7
 
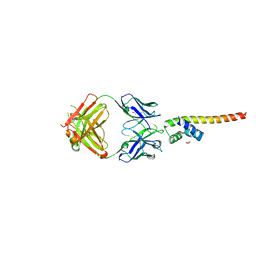 | | Fab 6D12 bound to ApoL1 NTD | | Descriptor: | Apolipoprotein L1, ETHANOL, Fab 6D12 heavy chain, ... | | Authors: | Ultsch, M, Kirchhofer, D. | | Deposit date: | 2021-01-15 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
7LFA
 
 | | Fab 3B6 bound to ApoL1 NTD | | Descriptor: | Apolipoprotein L1, CHLORIDE ION, Fab 3B6 heavy chain, ... | | Authors: | Ultsch, M, Kirchhofer, D. | | Deposit date: | 2021-01-15 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
7LHG
 
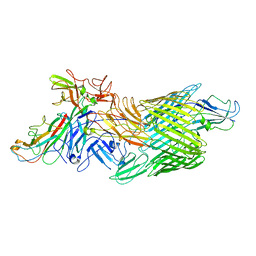 | | Cryo-EM structure of E. coli P pilus tip assembly intermediate PapC-PapD-PapK-PapG in the first conformation | | Descriptor: | Chaperone protein PapD, Fimbrial adapter PapK, P fimbria tip G-adhesin PapG-II, ... | | Authors: | Du, M, Yuan, Z, Werneburg, G, Henderson, N, Chauhan, H, Kovach, A, Zhao, G, Johl, J, Li, H, Thanassi, D. | | Deposit date: | 2021-01-23 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis.
Nat Commun, 12, 2021
|
|
6SPS
 
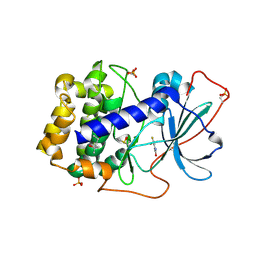 | | Crystal structure of cAMP-dependent protein kinase A (CHO PKA) in complex with 4-(trifluoromethyl)benzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(trifluoromethyl)benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Oebbeke, M, Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2019-09-02 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LHH
 
 | | Cryo-EM structure of E. coli P pilus tip assembly intermediate PapC-PapD-PapK-PapG in the second conformation | | Descriptor: | Chaperone protein PapD, Fimbrial adapter PapK, P fimbria tip G-adhesin PapG-II, ... | | Authors: | Du, M, Yuan, Z, Werneburg, G, Henderson, N, Chauhan, H, Kovach, A, Zhao, G, Johl, J, Li, H, Thanassi, D. | | Deposit date: | 2021-01-23 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis.
Nat Commun, 12, 2021
|
|
7LF8
 
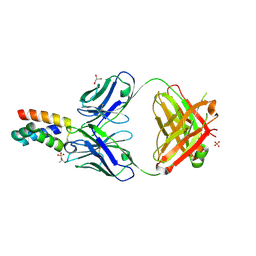 | | Fab 6D12 bound to ApoL2 NTD | | Descriptor: | Apolipoprotein L2, Fab 6D12 heavy chain, Fab 6D12 light chain, ... | | Authors: | Ultsch, M, Kirchhofer, D. | | Deposit date: | 2021-01-15 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
7LFD
 
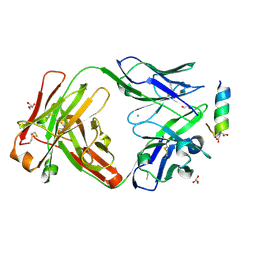 | | Fab 7D6 bound to ApoL1 BH3 like peptide | | Descriptor: | AMMONIUM ION, Apolipoprotein L1 BH3 like peptide, CITRATE ANION, ... | | Authors: | Ultsch, M, Kirchhofer, D. | | Deposit date: | 2021-01-16 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
6SVC
 
 | | Protein allostery of the WW domain at atomic resolution: apo structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7LHI
 
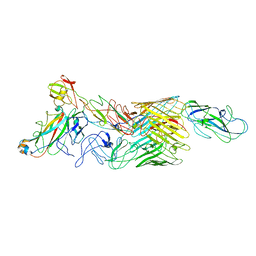 | | Cryo-EM structure of E. coli P pilus tip assembly intermediate PapC-PapD-PapK-PapF-PapG | | Descriptor: | Chaperone protein PapD, Fimbrial adapter PapK, Fimbrial protein, ... | | Authors: | Du, M, Yuan, Z, Werneburg, G, Henderson, N, Chauhan, H, Kovach, A, Zhao, G, Johl, J, Li, H, Thanassi, D. | | Deposit date: | 2021-01-23 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis.
Nat Commun, 12, 2021
|
|
6SSR
 
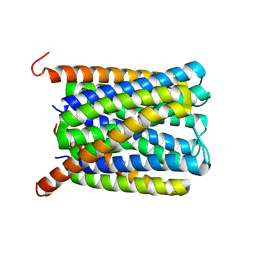 | |
1YIU
 
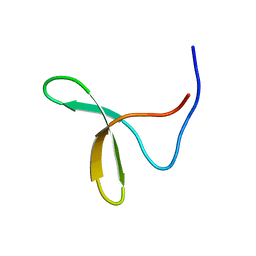 | | Itch E3 ubiquitin ligase WW3 domain | | Descriptor: | Itchy E3 ubiquitin protein ligase | | Authors: | Shaw, A.Z, Martin-Malpartida, P, Morales, B, Yraola, F, Royo, M, Macias, M.J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-08-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of either Ser16 or Thr30 does not disrupt the structure of the Itch E3 ubiquitin ligase third WW domain
Proteins, 60, 2005
|
|
7LHV
 
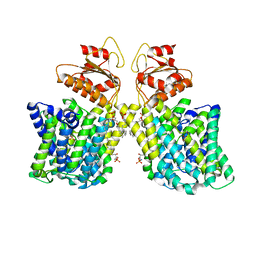 | | Structure of Arabidopsis thaliana sulfate transporter AtSULTR4;1 | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SULFATE ION, ... | | Authors: | Wang, L, Chen, K, Zhou, M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure and function of an Arabidopsis thaliana sulfate transporter.
Nat Commun, 12, 2021
|
|
6STE
 
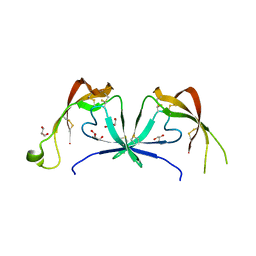 | |
6SPG
 
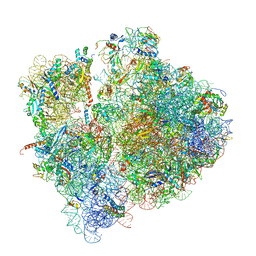 | | Pseudomonas aeruginosa 70s ribosome from a clinical isolate | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7LIB
 
 | |
7LGQ
 
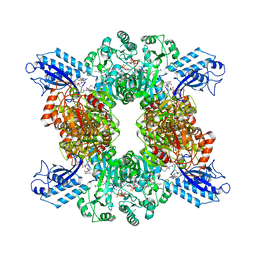 | | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ATP and 8x(Asp-Arg)-Asn | | Descriptor: | 8x(Asp-Arg)-Asn, ADENOSINE-5'-TRIPHOSPHATE, Cyanophycin synthase, ... | | Authors: | Sharon, I, Grogg, M, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and function of the amino acid polymerase cyanophycin synthetase.
Nat.Chem.Biol., 17, 2021
|
|
7LGJ
 
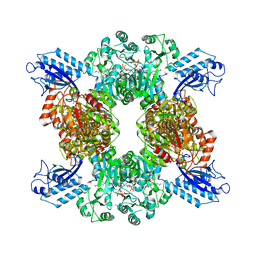 | | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ADPCP and 8x(Asp-Arg)-NH2 | | Descriptor: | 8x(Asp-Arg)-NH2, Cyanophycin synthase, MAGNESIUM ION, ... | | Authors: | Sharon, I, Grogg, M, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures and function of the amino acid polymerase cyanophycin synthetase.
Nat.Chem.Biol., 17, 2021
|
|
1Y2U
 
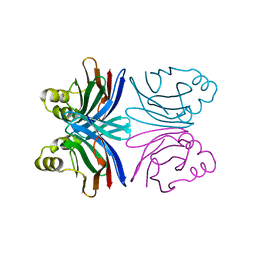 | | Crystal structure of the common edible mushroom (Agaricus bisporus) lectin in complex with Lacto-N-biose | | Descriptor: | beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin | | Authors: | Carrizo, M.E, Capaldi, S, Perduca, M, Irazoqui, F.J, Nores, G.A, Monaco, H.L. | | Deposit date: | 2004-11-23 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Antineoplastic Lectin of the Common Edible Mushroom (Agaricus bisporus) Has Two Binding Sites, Each Specific for a Different Configuration at a Single Epimeric Hydroxyl
J.Biol.Chem., 280, 2005
|
|
6SUD
 
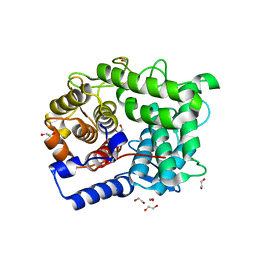 | | Structure of L320A mutant of Rex8A from Paenibacillus barcinonensis complexed with xylose. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Reducing-end xylose-releasing exo-oligoxylanase Rex8A, ... | | Authors: | Jimenez-Ortega, E, Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2019-09-13 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural analysis of the reducing-end xylose-releasing exo-oligoxylanase Rex8A from Paenibacillus barcinonensis BP-23 deciphers its molecular specificity.
Febs J., 287, 2020
|
|
6SWJ
 
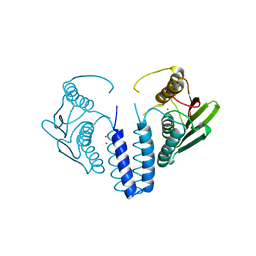 | | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus (with Pt) | | Descriptor: | Histidine kinase, PLATINUM (II) ION | | Authors: | Lansky, S, Shiradski, M, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2019-09-22 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus (with Pt)
To Be Published
|
|
7LPQ
 
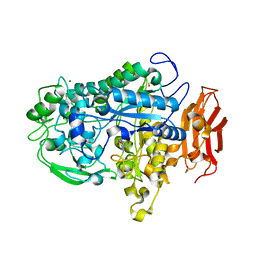 | | Crystal structure of Cryptococcus neoformans sterylglucosidase 1 with hit 9 | | Descriptor: | Cytoplasmic protein, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Pereira de Sa, N, Del Poeta, M, Airola, M.V. | | Deposit date: | 2021-02-12 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and inhibition of Cryptococcus neoformans sterylglucosidase to develop antifungal agents.
Nat Commun, 12, 2021
|
|
6SJR
 
 | | Methyltransferase of the MtgA N227A mutant from Desulfitobacterium hafniense in complex with tetrahydrofolate | | Descriptor: | GLYCEROL, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, Tetrahydromethanopterin S-methyltransferase | | Authors: | Badmann, T, Groll, M. | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures in Tetrahydrofolate Methylation in Desulfitobacterial Glycine Betaine Metabolism at Atomic Resolution.
Chembiochem, 21, 2020
|
|
1TML
 
 | |
