3ZX7
 
 | | Complex of lysenin with phosphocholine | | Descriptor: | LYSENIN, PHOSPHATE ION, PHOSPHOCHOLINE, ... | | Authors: | De Colibus, L, Sonnen, A.F.P, Morris, K.J, Siebert, C.A, Abrusci, P, Plitzko, J, Hodnik, V, Leippe, M, Volpi, E, Anderluh, G, Gilbert, R.J.C. | | Deposit date: | 2011-08-08 | | Release date: | 2012-09-19 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structures of Lysenin Reveal a Shared Evolutionary Origin for Pore-Forming Proteins and its Mode of Sphingomyelin Recognition.
Structure, 20, 2012
|
|
6TQZ
 
 | |
7QMS
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 2) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMU
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 4) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMV
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 5) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QTX
 
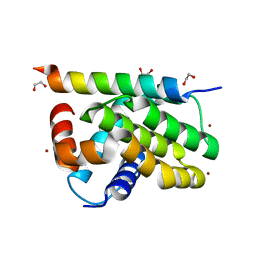 | | Kaposi sarcoma associated herpes virus (KSHV) encoded apoptosis inhibitor, KsBcl-2 in complex with Puma BH3 | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Bcl-2, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.11598849 Å) | | Cite: | Structural Insight into KsBcl-2 Mediated Apoptosis Inhibition by Kaposi Sarcoma Associated Herpes Virus.
Viruses, 14, 2022
|
|
6TT0
 
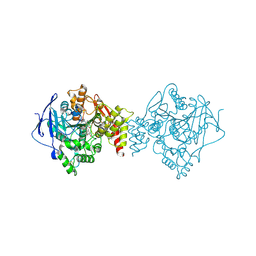 | | Crystal structure of a potent and reversible dual binding site Acetylcholinesterase chiral inhibitor | | Descriptor: | (1~{R},3~{S})-~{N}-(6,7-dimethoxy-2-oxidanylidene-chromen-3-yl)-3-[(phenylmethyl)amino]cyclohexane-1-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | de la Mora, E, Mangiatordi, G.F, Belviso, B.D, Caliandro, R, Colletier, J.P, Catto, M. | | Deposit date: | 2019-12-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.80003023 Å) | | Cite: | Chiral Separation, X-ray Structure, and Biological Evaluation of a Potent and Reversible Dual Binding Site AChE Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
6TT8
 
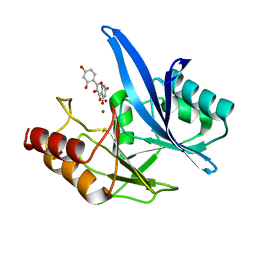 | | Haddock model of NDM-1/morin complex | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
3ZVJ
 
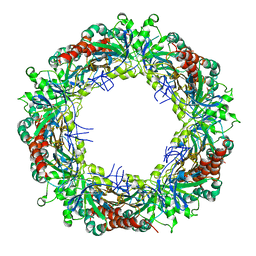 | | Crystal structure of high molecular weight (HMW) form of Peroxiredoxin I from Schistosoma mansoni | | Descriptor: | THIOREDOXIN PEROXIDASE | | Authors: | Saccoccia, F, Angelucci, F, Bellelli, A, Boumis, G, Brunori, M, Miele, A.E. | | Deposit date: | 2011-07-25 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Moonlighting by Different Stressors: Crystal Structure of the Chaperone Species of a 2-Cys Peroxiredoxin.
Structure, 20, 2012
|
|
7QTW
 
 | | Kaposi sarcoma associated herpes virus(KSHV) encoded apoptosis inhibitor, KsBcl-2 in complex with Bid BH3 | | Descriptor: | 1,2-ETHANEDIOL, BH3-interacting domain death agonist p15, Bcl-2 | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural Insight into KsBcl-2 Mediated Apoptosis Inhibition by Kaposi Sarcoma Associated Herpes Virus.
Viruses, 14, 2022
|
|
7QMW
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 6) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMX
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 7) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QN0
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 10) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3ZXJ
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HIAXHD3, ... | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXK
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIAXHD3, alpha-L-arabinofuranose-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6TTC
 
 | | Haddock model of NDM-1/myricetin complex | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
7QN2
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 12) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6TQV
 
 | |
9FEJ
 
 | | Structure of the RNA-dependent RNA polymerase P2 from the bacteriophage Phi8 | | Descriptor: | CALCIUM ION, RNA-directed RNA polymerase | | Authors: | Latimer-Smith, M, Salgado, P.S, Forsyth, I, Makeyev, E, Poranen, M, Stuart, D.I, Grimes, J.M, El Omari, K. | | Deposit date: | 2024-05-20 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase P2 from the cystovirus phi 8.
Sci Rep, 14, 2024
|
|
3ZYP
 
 | | Cellulose induced protein, Cip1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CIP1, ... | | Authors: | Jacobson, F, Karkehabadi, S, Hansson, H, Goedegebuur, F, Wallace, L, Mitchinson, C, Piens, K, Stals, I, Sandgren, M. | | Deposit date: | 2011-08-24 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of the Core Domain of a Cellulose Induced Protein (Cip1) from Hypocrea Jecorina, at 1.5 A Resolution.
Plos One, 8, 2013
|
|
3ZLI
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Vollmar, M, Gileadi, C, Shrestha, L, Goubin, S, Johansson, C, Krojer, T, Raynor, J.W, Bradley, A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Uty(Kdm6C) is a Male-Specific Nepsilon-Methyl Lysyl Demethylase.
J.Biol.Chem., 289, 2014
|
|
6TPS
 
 | | early intermediate RNA Polymerase I Pre-initiation complex - eiPIC | | Descriptor: | DNA foreign, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, ... | | Authors: | Pilsl, M, Engel, C. | | Deposit date: | 2019-12-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of RNA polymerase I pre-initiation complex formation and promoter melting.
Nat Commun, 11, 2020
|
|
3ZN8
 
 | | Structural Basis of Signal Sequence Surveillance and Selection by the SRP-SR Complex | | Descriptor: | 4.5 S RNA, DIPEPTIDYL AMINOPEPTIDASE B, MAGNESIUM ION, ... | | Authors: | von Loeffelholz, O, Knoops, K, Ariosa, A, Zhang, X, Karuppasamy, M, Huard, K, Schoehn, G, Berger, I, Shan, S.O, Schaffitzel, C. | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural Basis of Signal Sequence Surveillance and Selection by the Srp-Sr Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
6TTZ
 
 | | Structure of the ClpP:ADEP4-complex from Staphylococcus aureus (open state) | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, N-[(2S)-3-(3,5-difluorophenyl)-1-[[(3S,9S,13S,15R,19S,22S)-15,19-dimethyl-2,8,12,18,21-pentaoxo-11-oxa-1,7,17,20-tetrazatetracyclo[20.4.0.03,7.013,17]hexacosan-9-yl]amino]-1-oxopropan-2-yl]heptanamide | | Authors: | Malik, I.T, Pereira, R, Vielberg, M.-T, Mayer, C, Straetener, J, Thomy, D, Famulla, K, Castro, H.C, Sass, P, Groll, M, Broetz-Oesterheldt, H. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Characterisation of ClpP Mutations Conferring Resistance to Acyldepsipeptide Antibiotics in Firmicutes.
Chembiochem, 21, 2020
|
|
7QTV
 
 | | Beryllium fluoride form of the Na+,K+-ATPase (E2-BeFx) | | Descriptor: | 1-O-decanoyl-beta-D-tagatofuranosyl beta-D-allopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shahsavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | Deposit date: | 2022-01-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
