7QWJ
 
 | |
2YWF
 
 | | Crystal structure of GMPPNP-bound LepA from Aquifex aeolicus | | Descriptor: | GTP-binding protein lepA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Nishino, A, Nakayama-Ushikoshi, R, Hanawa-Suetsugu, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structures of GTP-binding protein LepA from Aquifex aeolicus
To be Published
|
|
5I69
 
 | | MBP-MamC magnetite-interaction component mutant-D70A | | Descriptor: | Maltose-binding periplasmic protein,Tightly bound bacterial magnetic particle protein,Maltose-binding periplasmic protein, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nudelman, H, Tercedor, C.V, Kolusheva, S, Gonzalez, T.P, Widdrat, M, Grimberg, N, Levi, H, Nelkenbaum, O, Davidove, G, Faivre, D, Jimenez-Lopez, C, Zarivach, R. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function studies of the magnetite-biomineralizing magnetosome-associated protein MamC.
J.Struct.Biol., 194, 2016
|
|
5I6I
 
 | | Crystal structure of a dBCCP-variant of Chaetomium thermophilum acetyl-CoA carboxylase | | Descriptor: | Acetyl-CoA carboxylase-like protein,Acetyl-CoA carboxylase-like protein,Acetyl-CoA carboxylase-like protein | | Authors: | Hunkeler, M, Stuttfeld, E, Hagmann, A, Imseng, S, Maier, T. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.4 Å) | | Cite: | The dynamic organization of fungal acetyl-CoA carboxylase.
Nat Commun, 7, 2016
|
|
5F64
 
 | | Putative positive transcription regulator (sensor EvgS) from Shigella flexneri | | Descriptor: | Positive transcription regulator EvgA | | Authors: | Nocek, B, Osipiuk, J, Mulligan, R, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Putative positive transcription regulator (sensor EvgS) from Shigella flexneri.
to be published
|
|
2YP1
 
 | | Crystallization of a 45 kDa peroxygenase- peroxidase from the mushroom Agrocybe aegerita and structure determination by SAD utilizing only the haem iron | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Piontek, K, Strittmatter, E, Ullrich, R, Plattner, D.A, Hofrichter, M. | | Deposit date: | 2012-10-29 | | Release date: | 2013-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis of Substrate Conversion in a New Aromatic Peroxygenase: P450 Functionality with Benefits
J.Biol.Chem., 288, 2013
|
|
8ITO
 
 | |
2YY8
 
 | |
2YQQ
 
 | | Solution structure of the zf-HIT domain in zinc finger HIT domain-containing protein 3 (TRIP-3) | | Descriptor: | ZINC ION, Zinc finger HIT domain-containing protein 3 | | Authors: | Suzuki, S, He, F, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zf-HIT domain in zinc finger HIT domain-containing protein 3 (TRIP-3)
To be Published
|
|
6XIW
 
 | | Cryo-EM structure of the sodium leak channel NALCN-FAM155A complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Chua, H.C, Noland, C.L, Weidling, C, Clairfeuille, T, Bahlke, O.O, Ameen, A.O, Li, Z.R, Arthur, C.P, Ciferri, C, Pless, S.A, Payandeh, J. | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-29 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human sodium leak channel NALCN.
Nature, 587, 2020
|
|
5I72
 
 | | Crystal structure of the oligomeric form of the Lassa virus matrix protein Z | | Descriptor: | RING finger protein Z, ZINC ION | | Authors: | Hastie, K, Zandonatti, M, Liu, T, Li, S, Woods Jr, V, Saphire, E.O. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Oligomeric Form of Lassa Virus Matrix Protein Z.
J.Virol., 90, 2016
|
|
3TLS
 
 | | The GLIC pentameric Ligand-Gated Ion Channel E19'P mutant in a locally-closed conformation (LC2 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5F5V
 
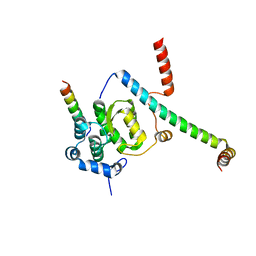 | | Crystal structure of the Snu23-Prp38-MFAP1(217-296) complex of Chaetomium thermophilum | | Descriptor: | Prp38, Putative uncharacterized protein, Zinc finger domain-containing protein | | Authors: | Ulrich, A.K.C, Seeger, M, Bartlick, N, Wahl, M.C. | | Deposit date: | 2015-12-04 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Scaffolding in the Spliceosome via Single alpha Helices.
Structure, 24, 2016
|
|
5EV7
 
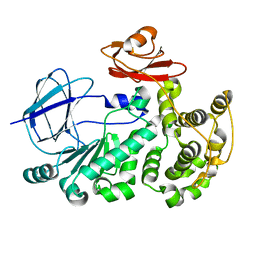 | | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames | | Descriptor: | Conserved domain protein | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames.
To Be Published
|
|
5IAT
 
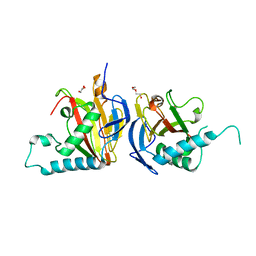 | |
5I7N
 
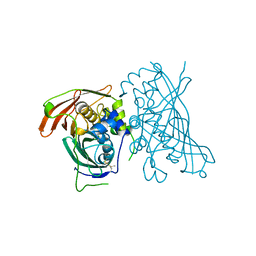 | | MaoC-like dehydratase | | Descriptor: | MaoC-like dehydratase | | Authors: | Blaise, M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deletion of a dehydratase important for intracellular growth and cording renders rough Mycobacterium abscessus avirulent.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7QW4
 
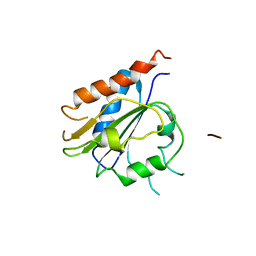 | | Pden_5119 protein | | Descriptor: | NADPH-dependent FMN reductase | | Authors: | Kryl, M, Sedlacek, V. | | Deposit date: | 2022-01-24 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Insight into Catalysis by the Flavin-Dependent NADH Oxidase (Pden_5119) of Paracoccus denitrificans .
Int J Mol Sci, 24, 2023
|
|
5IB5
 
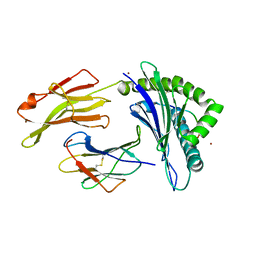 | | Crystal structure of HLA-B*27:09 complexed with the self-peptide pVIPR and Copper | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
5EWF
 
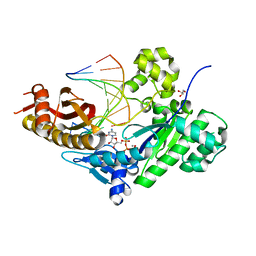 | | Ternary complex of human DNA polymerase eta inserting rCTP opposite an 8-Oxodeoxyguanosine Lesion | | Descriptor: | CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Su, Y, Egli, M, Guengerich, F.P. | | Deposit date: | 2015-11-20 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Mechanism of Ribonucleotide Incorporation by Human DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
8IU6
 
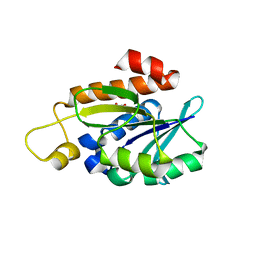 | | Crystal structure of peptidyl-tRNA hydrolase mutant from Enterococcus faecium | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Pandey, R, Tripathi, S, Lanka, A.K, Zohib, M, Pal, R.K, Arora, A. | | Deposit date: | 2023-03-23 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase mutant from Enterococcus faecium
To Be Published
|
|
2YTZ
 
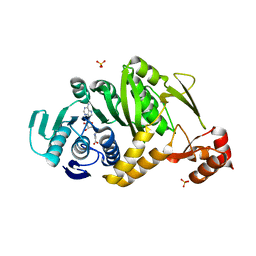 | | Complex structure of Trm1 from Pyrococcus horikoshii with S-adenosyl-L-Homocystein in the orthorhombic crystal-lattice | | Descriptor: | N(2),N(2)-dimethylguanosine tRNA methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Ihsanawati, Shirouzu, M, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of tRNA N(2),N(2)-Guanosine Dimethyltransferase Trm1 from Pyrococcus horikoshii
J.Mol.Biol., 383, 2008
|
|
8J7S
 
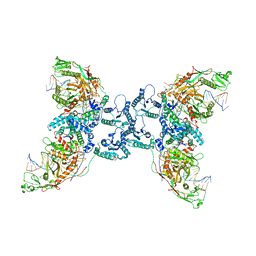 | | Structure of the SPARTA complex | | Descriptor: | DNA (5'-D(P*TP*AP*AP*TP*AP*GP*AP*TP*TP*AP*GP*AP*GP*CP*CP*GP*TP*CP*AP*AP*TP*AP*GP*A)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*A)-3'), ... | | Authors: | Guo, M, Zhu, Y, Lin, Z, Huang, Z. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the ssDNA-activated SPARTA complex.
Cell Res., 33, 2023
|
|
5IB2
 
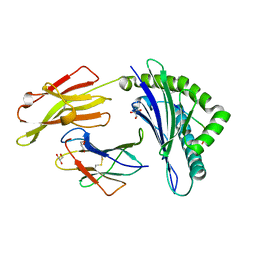 | | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
6XKU
 
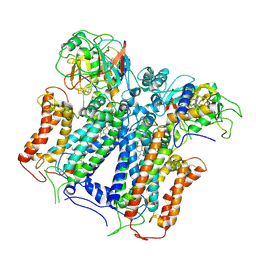 | | R. capsulatus cyt bc1 with one FeS protein in b position and one in c position (CIII2 b-c) | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
5EWQ
 
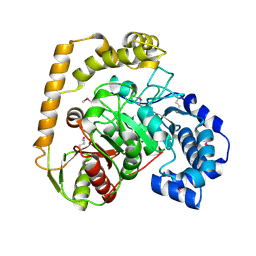 | | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames | | Descriptor: | ACETATE ION, Amidase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames
To Be Published
|
|
