3W5M
 
 | | Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Putative rhamnosidase | | Authors: | Fujimoto, Z, Jackson, A, Michikawa, M, Maehara, T, Momma, M, Henrissat, B.F, Gilbert, H.J, Kaneko, S. | | Deposit date: | 2013-01-31 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a Streptomyces avermitilis alpha-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement.
J.Biol.Chem., 288, 2013
|
|
6NW0
 
 | | Crystal Structure Desulfovibrio desulfuricans Nickel-Substituted Rubredoxin | | Descriptor: | NICKEL (II) ION, Rubredoxin | | Authors: | Slater, J.W, Marguet, S.C, Gray, M.E, Sotomayor, M, Shafaat, H.S. | | Deposit date: | 2019-02-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Power of the Secondary Sphere: Modulating Hydrogenase Activity in Nickel-Substituted Rubredoxin
Acs Catalysis, 2019
|
|
8CNY
 
 | | Structure of Enterovirus A71 3C protease | | Descriptor: | Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of EV D68 3C protease - to be published
To Be Published
|
|
7JSR
 
 | | Crystal structure of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Lazaro, M, Melero, R, Huet, C, Lopez-Alonso, J.P, Delgado, S, Dodu, A, Bruch, E.M, Abriata, L.A, Alzari, P.M, Valle, M, Lisa, M.N. | | Deposit date: | 2020-08-15 | | Release date: | 2021-06-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (6.27 Å) | | Cite: | 3D architecture and structural flexibility revealed in the subfamily of large glutamate dehydrogenases by a mycobacterial enzyme.
Commun Biol, 4, 2021
|
|
6NC5
 
 | | Cronobacter sakazakii (Enterobacter sakazakii) Metallo-beta-lactamse HARLDQ motif | | Descriptor: | ACETATE ION, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-12-10 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.074 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse.
Protein Cell, 11, 2020
|
|
6NIA
 
 | |
1UHG
 
 | | Crystal Structure of S-Ovalbumin At 1.9 Angstrom Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ovalbumin, ... | | Authors: | Yamasaki, M, Takahashi, N, Hirose, M. | | Deposit date: | 2003-07-03 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of S-ovalbumin as a Non-loop-inserted Thermostabilized Serpin Form
J.Biol.Chem., 278, 2003
|
|
6NJ1
 
 | |
2YR4
 
 | | Crystal structure of L-phenylalanine oxiase from Psuedomonas sp. P-501 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pro-enzyme of L-phenylalanine oxidase, SULFATE ION | | Authors: | Ida, K, Kurabayashi, M, Suguro, M, Suzuki, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
J.Biol.Chem., 283, 2008
|
|
7STF
 
 | | Structure of KRAS G12V/HLA-A*03:01 in complex with antibody fragment V2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Wright, K.M, Gabelli, S.B, Miller, M. | | Deposit date: | 2021-11-12 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Hydrophobic interactions dominate the recognition of a KRAS G12V neoantigen.
Nat Commun, 14, 2023
|
|
3DM8
 
 | | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris | | Descriptor: | DODECYL NONA ETHYLENE GLYCOL ETHER, uncharacterized protein RPA4348 | | Authors: | Cymborowski, M, Chruszcz, M, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris
To be Published
|
|
6NLN
 
 | | 1.60 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor (analog 16) | | Descriptor: | 4-{[3-(3-hydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
3Q5I
 
 | | Crystal Structure of PBANKA_031420 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, Panico, E, Crombet, L, Cossar, D, Hassani, A, Vedadi, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Neculai, A.M, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of CDPK1 from Plasmodium Bergheii, PBANKA_031420
To be Published
|
|
5HCB
 
 | | Globular Domain of the Entamoeba histolytica calreticulin in complex with glucose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calreticulin, ... | | Authors: | Moreau, C.P, Cioci, G, Ianello, M, Laffly, E, Chouquet, A, Ferreira, A, Thielens, N.M, Gaboriaud, C. | | Deposit date: | 2016-01-04 | | Release date: | 2016-08-31 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of parasite calreticulins provide insights into their flexibility and dual carbohydrate/peptide-binding properties.
IUCrJ, 3, 2016
|
|
6GLZ
 
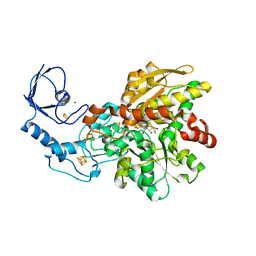 | | [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant C299D | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, Iron hydrogenase 1, ... | | Authors: | Duan, J, Esselborn, J, Hofmann, E, Winkler, M, Happe, T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystallographic and spectroscopic assignment of the proton transfer pathway in [FeFe]-hydrogenases.
Nat Commun, 9, 2018
|
|
7TA8
 
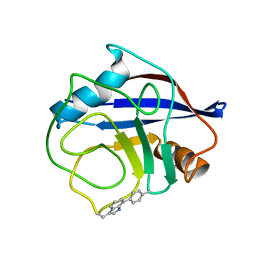 | | NMR structure of crosslinked cyclophilin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Lu, M, Toptygin, D, Xiang, Y, Shi, Y, Schwieters, C.D, Lipinski, E.C, Ahn, J, Byeon, I.-J.L, Gronenborn, A.M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Magic of Linking Rings: Discovery of a Unique Photoinduced Fluorescent Protein Crosslink.
J.Am.Chem.Soc., 144, 2022
|
|
6NRV
 
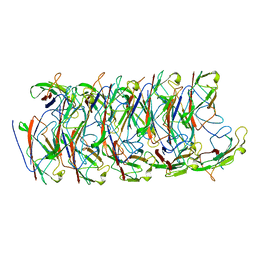 | |
7T4V
 
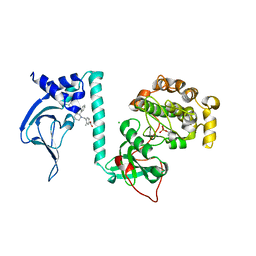 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-methylbenzoic acid, CHLORIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
7T4W
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | (2R)-2-({(2S,3S)-1-[(1H-benzimidazol-2-yl)methyl]-2-phenylpiperidin-3-yl}oxy)-2-(3,5-dichlorophenyl)ethan-1-ol, CHLORIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
6HLT
 
 | |
6NIM
 
 | | Trypanosoma cruzi - BDF2, TcCLB.506553.20, solved with bromosporine | | Descriptor: | Bromodomain factor 2 protein, Bromosporine, SULFATE ION, ... | | Authors: | Lin, Y.H, Dong, A, Tempel, W, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Vedadi, M, Harding, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-29 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Trypanosoma cruzi - BDF2, TcCLB.506553.20, solved with bromosporine
to be published
|
|
6HLV
 
 | |
6HMF
 
 | | D-family DNA polymerase - DP1 subunit (3'-5' proof-reading exonuclease) H451 proof-reading deficient variant | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Raia, P, Delarue, M, Sauguet, L. | | Deposit date: | 2018-09-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases.
PLoS Biol., 17, 2019
|
|
7T4U
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 3-amino-4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)benzoic acid, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
6NLG
 
 | | 1.50 A resolution structure of BfrB (C89S/K96C) from Pseudomonas aeruginosa in complex with a small molecule fragment (analog 1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-hydroxy-1H-isoindole-1,3(2H)-dione, Bacterioferritin, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
