2CMC
 
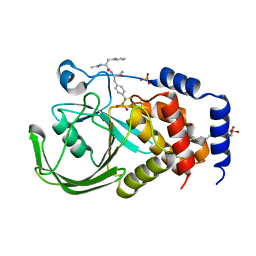 | | Structural Basis for Inhibition of Protein Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics | | Descriptor: | N-ACETYL-L-PHENYLALANYL-4-[DIFLUORO(PHOSPHONO)METHYL]-L-PHENYLALANINAMIDE, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1, ... | | Authors: | Ala, P.J, Gonneville, L, Hillman, M.C, Becker-Pasha, M, Wei, M, Reid, B.G, Klabe, R, Yue, E.W, Wayland, B, Douty, B, Combs, A.P, Polam, P, Wasserman, Z, Bower, M, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-04 | | Release date: | 2006-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Inhibition of Protein-Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics.
J.Biol.Chem., 281, 2006
|
|
2QC8
 
 | | Crystal structure of human glutamine synthetase in complex with ADP and methionine sulfoximine phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Glutamine synthetase, ... | | Authors: | Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hogbom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-19 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design.
J.Mol.Biol., 375, 2008
|
|
7COK
 
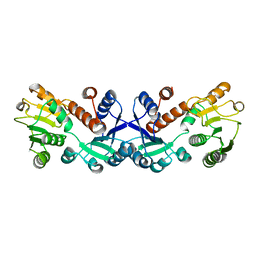 | | Crystal structure of ligand-free form of 5-ketofructose reductase of Gluconobacter sp. strain CHM43 | | Descriptor: | 5-ketofructose reductase | | Authors: | Noda, S, Hodoya, Y, Nguyen, T.M, Kataoka, N, Adachi, O, Matsutani, M, Matsushita, K, Yakushi, T, Goto, M. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 5-Ketofructose Reductase of Gluconobacter sp. Strain CHM43 Is a Novel Class in the Shikimate Dehydrogenase Family.
J.Bacteriol., 203, 2021
|
|
5G4H
 
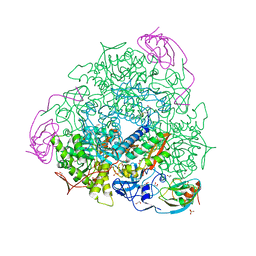 | | 1.50 A resolution catechol (1,2-dihydroxybenzene) inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, CATECHOL, HYDROXIDE ION, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2016-05-13 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inactivation of Urease by Catechol: Kinetics and Structure.
J.Inorg.Biochem., 166, 2016
|
|
7COL
 
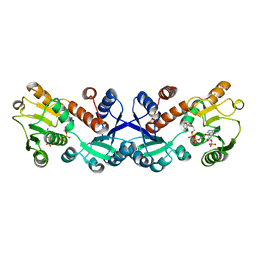 | | Crystal structure of 5-ketofructose reductase complexed with NADPH | | Descriptor: | 5-ketofructose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hodoya, Y, Noda, S, Nguyen, T.M, Kataoka, N, Adachi, O, Matsutani, M, Matsushita, K, Yakushi, T, Goto, M. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The 5-Ketofructose Reductase of Gluconobacter sp. Strain CHM43 Is a Novel Class in the Shikimate Dehydrogenase Family.
J.Bacteriol., 203, 2021
|
|
3MRE
 
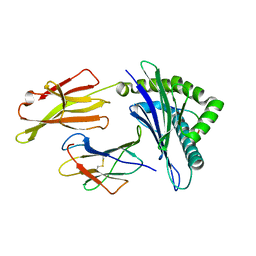 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with EBV bmlf1-280-288 nonapeptide | | Descriptor: | 9-meric peptide from mRNA export factor EB2, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
7CSM
 
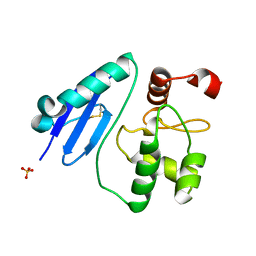 | | Crystal structure of Sulfurisphaera tokodaii O6-methylguanine methyltransferase C120S variant | | Descriptor: | Methylated-DNA--protein-cysteine methyltransferase, SULFATE ION | | Authors: | Kikuchi, M, Yamauchi, T, Iizuka, Y, Tsunoda, M. | | Deposit date: | 2020-08-15 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6JYQ
 
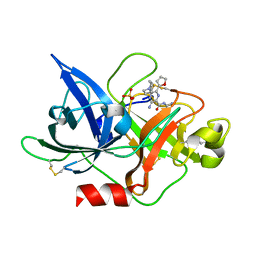 | | Crystal structure of uPA_H99Y in complex with 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(furan-2-yl)pyrazine-2-carboxamide | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(furan-2-yl)pyrazine-2-carboxamide, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2019-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | H99Y-6F-HMA-pH7
To Be Published
|
|
6UCI
 
 | | Transcription factor DeltaFosB bZIP domain self-assembly, oxidized form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein fosB | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2019-09-16 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Self-assembly of the bZIP transcription factor Delta FosB.
Curr Res Struct Biol, 2, 2020
|
|
6UCL
 
 | |
2VYZ
 
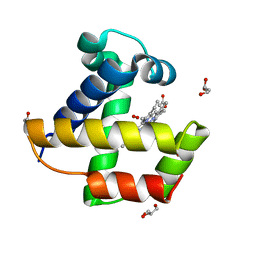 | | Mutant Ala55Phe of Cerebratulus lacteus mini-hemoglobin | | Descriptor: | ACETATE ION, GLYCEROL, NEURAL HEMOGLOBIN, ... | | Authors: | Salter, M.D, Nienhaus, K, Nienhaus, G.U, Dewilde, S, Moens, L, Pesce, A, Nardini, M, Bolognesi, M, Olson, J.S. | | Deposit date: | 2008-07-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Apolar Channel in Cerebratulus Lacteus Hemoglobin is the Route for O2 Entry and Exit.
J.Biol.Chem., 283, 2008
|
|
1J02
 
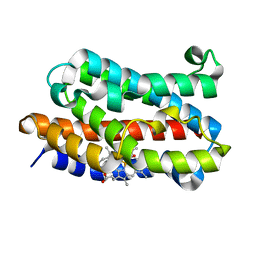 | | Crystal Structure of Rat Heme Oxygenase-1-Heme Bound to NO | | Descriptor: | HEME OXYGENASE 1, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Fukuyama, K. | | Deposit date: | 2002-10-28 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
3NBT
 
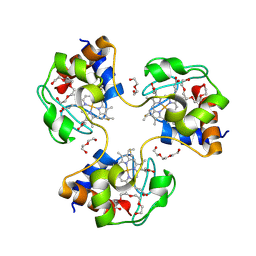 | | Crystal structure of trimeric cytochrome c from horse heart | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | Deposit date: | 2010-06-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2RCQ
 
 | | Crystal structure of human apo Cellular Retinol Binding Protein II (CRBP-II) | | Descriptor: | L(+)-TARTARIC ACID, Retinol-binding protein II, cellular, ... | | Authors: | Monaco, H.L, Capaldi, S, Perduca, M. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of human cellular retinol-binding protein II to 1.2 A resolution.
Proteins, 70, 2007
|
|
5IE0
 
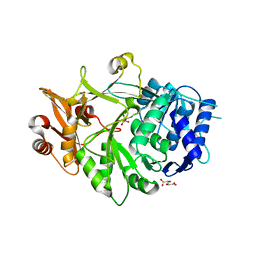 | | Crystal structure of a plant enzyme | | Descriptor: | Oxalate--CoA ligase, S,R MESO-TARTARIC ACID | | Authors: | Ran, M.R, Li, M, Chang, W.R. | | Deposit date: | 2016-02-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Oxalyl-CoA Synthetase Essential for Oxalate Degradation
Mol Plant, 9, 2016
|
|
3NHW
 
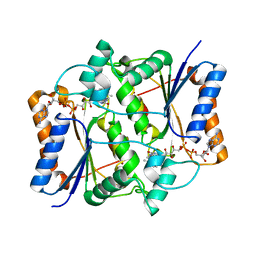 | |
7GRZ
 
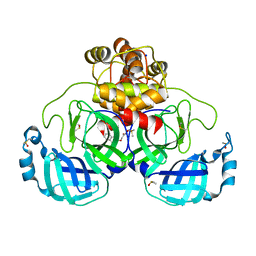 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-22 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N,N-dimethyl-2-[(naphthalen-2-yl)oxy]acetamide, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6D8Q
 
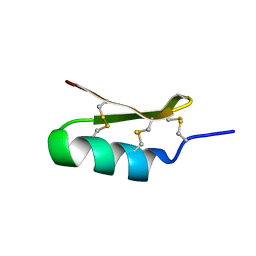 | |
7GS5
 
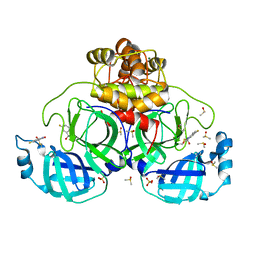 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-28 | | Descriptor: | (3R)-5-fluoro-3-hydroxy-1,3-dihydro-2H-indol-2-one, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS3
 
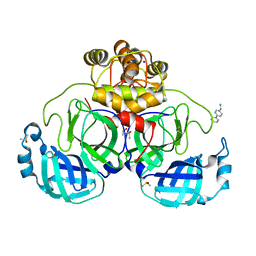 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-26 | | Descriptor: | (6-phenylpyridin-3-yl)methanamine, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6D8Y
 
 | |
7GS2
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-25 | | Descriptor: | 3-(pyridin-3-yl)benzoic acid, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRH
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-4 | | Descriptor: | 3C-like proteinase nsp5, 5-chloropyridin-3-ol, DIMETHYL SULFOXIDE, ... | | Authors: | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6D8R
 
 | |
6D93
 
 | |
