6ZE1
 
 | | human NBD1 of CFTR in complex with nanobody G11a | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Garcia-Pino, A, Govaerts, C, Scholl, D, Sigoillot, M. | | Deposit date: | 2020-06-15 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | A topological switch in CFTR modulates channel activity and sensitivity to unfolding.
Nat.Chem.Biol., 17, 2021
|
|
7MBL
 
 | | Crystal structure of Equine Serum Albumin in complex with Cobalt (II) | | Descriptor: | COBALT (II) ION, SULFATE ION, Serum albumin | | Authors: | Shabalin, I.G, Czub, M.P, Handing, K.B, Cymborowski, M.T, Grabowski, M, Cooper, D.R, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-03-31 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical characterisation of Co 2+ -binding sites on serum albumins and their interplay with fatty acids.
Chem Sci, 14, 2023
|
|
6ZHS
 
 | | Uba1 bound to two E2 (Ubc13) molecules | | Descriptor: | GLYCEROL, SULFATE ION, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system.
To Be Published
|
|
6ZHT
 
 | | Uba1-Ubc13 disulfide mediated complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Schaefer, A, Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system.
To Be Published
|
|
6ZHU
 
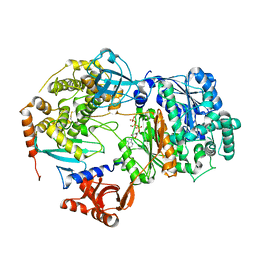 | | Yeast Uba1 in complex with Ubc3 and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system
To Be Published
|
|
6ERZ
 
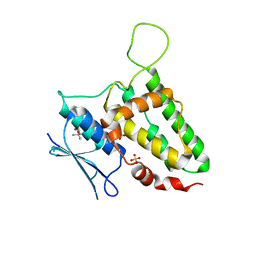 | | The crystal structure of mouse chloride intracellular channel protein 6 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chloride intracellular channel protein 6, SULFATE ION | | Authors: | Ferofontov, A, Giladi, M, Haitin, Y. | | Deposit date: | 2017-10-19 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Inherent flexibility of CLIC6 revealed by crystallographic and solution studies.
Sci Rep, 8, 2018
|
|
1QYU
 
 | |
4MMK
 
 | | Q8A Hfq from Pseudomonas aeruginosa | | Descriptor: | POTASSIUM ION, Protein hfq, SODIUM ION, ... | | Authors: | Murina, V.N, Filimonov, V.V, Melnik, B.S, Uhlein, M, Mueller, U, Weiss, M, Nikulin, A.D. | | Deposit date: | 2013-09-09 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Effect of conserved intersubunit amino Acid substitutions on hfq protein structure and stability.
Biochemistry Mosc., 79, 2014
|
|
1QZ3
 
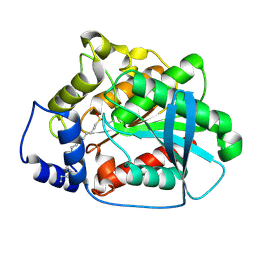 | | CRYSTAL STRUCTURE OF MUTANT M211S/R215L OF CARBOXYLESTERASE EST2 COMPLEXED WITH HEXADECANESULFONATE | | Descriptor: | 1-HEXADECANOSULFONIC ACID, CARBOXYLESTERASE EST2 | | Authors: | De Simone, G, Mandrich, L, Menchise, V, Giordano, V, Febbraio, F, Rossi, M, Pedone, C, Manco, G. | | Deposit date: | 2003-09-15 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A substrate-induced switch in the reaction mechanism of a thermophilic esterase: kinetic evidences and structural basis.
J.Biol.Chem., 279, 2004
|
|
6ZMC
 
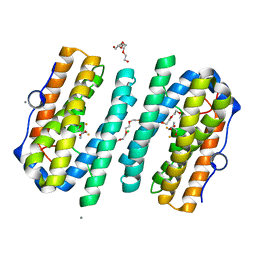 | | Structure of the tRNA-Monooxygenase enzyme MiaE frozen under 2000 bar using the high pressure freezing method | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ARGON, CALCIUM ION, ... | | Authors: | Carpentier, P, Atta, M. | | Deposit date: | 2020-07-02 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.
Nucleic Acids Res., 48, 2020
|
|
6EFK
 
 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated HSP70 peptide | | Descriptor: | ACE-ILE-GLU-GLU-VAL-ASP, E3 ubiquitin-protein ligase CHIP, SODIUM ION | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2018-08-16 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
6TVV
 
 | | Crystal structure of 3'-5' RecJ exonuclease from M. Jannaschii | | Descriptor: | GLYCEROL, MANGANESE (II) ION, MjaRecJ | | Authors: | De March, M, Medagli, B, Krastanova, I, Saha, I, Pisani, F, Onesti, S. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of 3'-5' RecJ exonuclease from M. Jannaschii
To Be Published
|
|
6EIC
 
 | | Crystal structure of Rv0183, a Monoglyceride Lipase from Mycobacterium Tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Mycobacterium Tuberculosis Monoglyceride Lipase, NITRATE ION, ... | | Authors: | Aschauer, P, Pavkov-Keller, T, Oberer, M. | | Deposit date: | 2017-09-19 | | Release date: | 2018-06-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of monoacylglycerol lipase from M. tuberculosis reveals the basis for specific inhibition.
Sci Rep, 8, 2018
|
|
1QKN
 
 | |
6ZO1
 
 | | 1.61 A resolution 3,5-dimethylcatechol (3,5-dimethylbenzene-1,2-diol) inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of Urease, a Ni-Enzyme: The Reactivity of a Key Thiol With Mono- and Di-Substituted Catechols Elucidated by Kinetic, Structural, and Theoretical Studies.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6U42
 
 | | Natively decorated ciliary doublet microtubule | | Descriptor: | DC1, DC2, DC3, ... | | Authors: | Ma, M, Stoyanova, M, Rademacher, G, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2019-08-22 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the Decorated Ciliary Doublet Microtubule.
Cell, 179, 2019
|
|
6ZTJ
 
 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (38 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTP
 
 | | E. coli 70S-RNAP expressome complex in uncoupled state 6 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
1QKM
 
 | |
6E7K
 
 | |
6EHJ
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and peptide bound | | Descriptor: | ASPARAGINE, COENZYME A, GLYCEROL, ... | | Authors: | Perez-Dorado, I, Ritzefeld, M, Tate, E.W. | | Deposit date: | 2017-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
6ECM
 
 | |
6UMW
 
 | | Crystal structure of hEphB1 bound with chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, Ephrin type-B receptor 1 | | Authors: | Ahmed, M, Wang, P, Sadek, H. | | Deposit date: | 2019-10-10 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6EMW
 
 | | Structure of S.aureus ClpC in complex with MecA | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit, ATP-dependent Clp protease ATP-binding subunit ClpC, Adapter protein MecA, ... | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6EO9
 
 | | Crystal structure of thrombin in complex with a novel glucose-conjugated potent inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, Hirudin variant-2, N-(2-{[5-(5-chlorothiophen-2-yl)-1,2-oxazol-3-yl]methoxy}-6-{3-[(2,3,4,6-tetra-O-acetyl-beta-D-glucopyranosyl)oxy]propoxy}phenyl)-1-(propan-2-yl)piperidine-4-carboxamide, ... | | Authors: | Belviso, B.D, Caliandro, R, Aresta, B.M, De Candia, M, Altomare, C.D. | | Deposit date: | 2017-10-09 | | Release date: | 2017-12-13 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How a beta-D-glucoside side chain enhances binding affinity to thrombin of inhibitors bearing 2-chlorothiophene as P1 moiety: crystallography, fragment deconstruction study, and evaluation of antithrombotic properties.
J. Med. Chem., 57, 2014
|
|
