6ZJE
 
 | | Crystal structure of human adenylate kinase 3, AK3, in complex with inhibitor Ap5A | | Descriptor: | BIS(ADENOSINE)-5'-PENTAPHOSPHATE, CHLORIDE ION, GTP:AMP phosphotransferase AK3, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Basis for GTP versus ATP Selectivity in the NMP Kinase AK3.
Biochemistry, 59, 2020
|
|
7Q92
 
 | | Crystal Structure of Agrobacterium tumefaciens NADQ, ATP complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, NADQ transcription factor, POTASSIUM ION, ... | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
5WOP
 
 | | High Resolution Structure of Mutant CA09-PB2cap | | Descriptor: | GLYCEROL, Polymerase PB2 | | Authors: | Constantinides, A.E, Gumpper, R.H, Severin, C, Luo, M. | | Deposit date: | 2017-08-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | High-resolution structure of the Influenza A virus PB2cap binding domain illuminates the changes induced by ligand binding.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7QBR
 
 | | Human butyrylcholinesterase in complex with (Z)-N-tert-butyl-1-(8-(3-(4-(prop-2-yn-1-yl)piperazin-1-yl)propoxy)quinolin-2-yl)methanimine oxide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Denic, M, Chioua, M, Knez, D, Gobec, S, Nachon, F, Marco-Contelles, J.L, Brazzolotto, X. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | 8-Hydroxyquinolylnitrones as multifunctional ligands for the therapy of neurodegenerative diseases.
Acta Pharm Sin B, 13, 2023
|
|
7QBQ
 
 | | Human butyrylcholinesterase in complex with (Z)-N-benzyl-1-(8-hydroxyquinolin-2-yl)methanimine oxide | | Descriptor: | 1-(8-oxidanylquinolin-2-yl)-N-(phenylmethyl)methanimine oxide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Denic, M, Chioua, M, Knez, D, Gobec, S, Nachon, F, Marco-Contelles, J.L, Brazzolotto, X. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | 8-Hydroxyquinolylnitrones as multifunctional ligands for the therapy of neurodegenerative diseases.
Acta Pharm Sin B, 13, 2023
|
|
6Z1I
 
 | |
6Z1O
 
 | |
6Z2B
 
 | |
1QUA
 
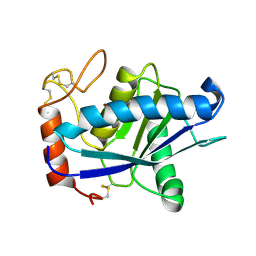 | | CRYSTAL STRUCTURE OF ACUTOLYSIN-C, A HEMORRHAGIC TOXIN FROM THE SNAKE VENOM OF AGKISTRODON ACUTUS, AT 2.2 A RESOLUTION | | Descriptor: | ACUTOLYSIN-C, ZINC ION | | Authors: | Niu, L, Teng, M, Zhu, X. | | Deposit date: | 1999-06-30 | | Release date: | 2000-07-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of acutolysin-C, a haemorrhagic toxin from the venom of Agkistrodon acutus, providing further evidence for the mechanism of the pH-dependent proteolytic reaction of zinc metalloproteinases.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7QFJ
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II (aa 194-362) | | Descriptor: | SlpX | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C, Baek, M, Read, R, Baker, D. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6Z1C
 
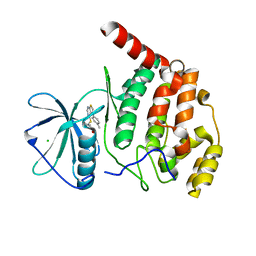 | | Crystal structure of Arabidopsis thaliana CK2-alpha-1 in complex with TTP-22 | | Descriptor: | 3-[5-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]sulfanylpropanoic acid, CHLORIDE ION, Casein kinase II subunit alpha-1 | | Authors: | Demulder, M, Loris, R, De Veylder, L. | | Deposit date: | 2020-05-13 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arabidopsis casein kinase 2 triggers stem cell exhaustion under Al toxicity and phosphate deficiency through activating the DNA damage response pathway.
Plant Cell, 33, 2021
|
|
6ZC2
 
 | | Crystal structure of RahU protein in complex with TRIS molecule | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RahU protein | | Authors: | Podobnik, M, Anderluh, G, Lenarcic, T. | | Deposit date: | 2020-06-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structure of RahU, an aegerolysin protein from the human pathogen Pseudomonas aeruginosa, and its interaction with membrane ceramide phosphorylethanolamine.
Sci Rep, 11, 2021
|
|
6ZC1
 
 | |
6TH8
 
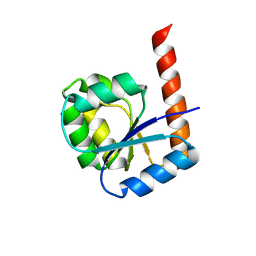 | |
6ZEY
 
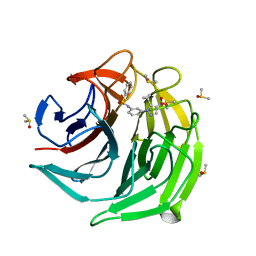 | |
6ZF1
 
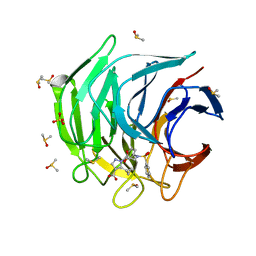 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[2-hydroxy-2-oxoethyl(phenylsulfonyl)amino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6EO8
 
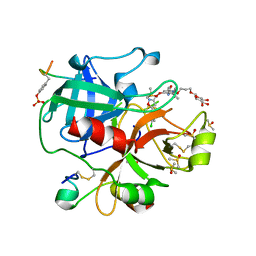 | | Crystal structure of thrombin in complex with a novel glucose-conjugated potent inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, Hirudin variant-2, N-(2-{[5-(5-chlorothiophen-2-yl)-1,2-oxazol-3-yl]methoxy}-6-[3-(beta-D-glucopyranosyloxy)propoxy]phenyl)-1-(propan-2-yl)piperidine-4-carboxamide, ... | | Authors: | Belviso, B.D, Caliandro, R, Aresta, B.M, De Candia, M, Altomare, C.D. | | Deposit date: | 2017-10-09 | | Release date: | 2017-12-13 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | How a beta-D-glucoside side chain enhances binding affinity to thrombin of inhibitors bearing 2-chlorothiophene as P1 moiety: crystallography, fragment deconstruction study, and evaluation of antithrombotic properties.
J. Med. Chem., 57, 2014
|
|
6ZF2
 
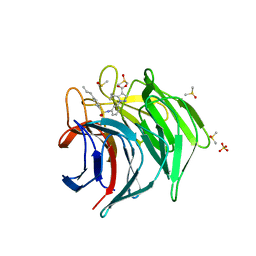 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[(4-propylphenyl)sulfonylamino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
7LBC
 
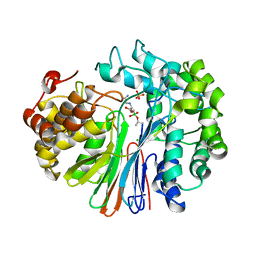 | | Structure of human GGT1 in complex with Lnt2-65 compound | | Descriptor: | (2R)-2-[(2-aminoethyl)amino]-4-boronobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2021-01-07 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Design and evaluation of novel analogs of 2-amino-4-boronobutanoic acid (ABBA) as inhibitors of human gamma-glutamyl transpeptidase.
Bioorg.Med.Chem., 73, 2022
|
|
6ZF6
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 1-[3-[2-hydroxy-2-oxoethyl(phenylsulfonyl)amino]phenyl]-5-[(1~{S},2~{S})-2-phenylcyclopropyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF0
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[(2,3,5,6-tetramethylphenyl)sulfonylamino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF3
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, SULFATE ION, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF8
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 1-[3-[2-hydroxy-2-oxoethyl-(3-methoxyphenyl)sulfonyl-amino]phenyl]-5-[(1~{S},2~{S})-2-phenylcyclopropyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6TI6
 
 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6ZEW
 
 | | Keap1 kelch domain bound to a small molecule fragment | | Descriptor: | 7-methoxy-1~{H}-benzotriazole, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
