4RFO
 
 | | Crystal structure of the ADCC-Potent Antibody N60-I3 Fab in complex with HIV-1 Clade A/E gp120 and M48u1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade A/E gp120, N60-i3 Fab heavy chain, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
8GRT
 
 | | Small Dipeptide Analogues developed by Co-crystal Structure of Stenotrophomonas maltophilia Dipeptidyl Peptidase 7 | | Descriptor: | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID, Dipeptidyl-peptidase, TYROSINE | | Authors: | Yasumitsu, S, Koushi, H, Akihiro, N, Yoshiyuki, Y, Wataru, O, Mizuki, S, Saori, R, Nobutada, T, Anna, M, Keiko, H, Tsuda, Y. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Small Dipeptide Analogues Generated by Co-crystal Structure of Bacterial Dipeptidyl Peptidase 7 to Defeat Stenotrophomonas maltophilia
To Be Published
|
|
6LJG
 
 | | Crassostrea gigas ferritin mutant-D119G | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Li, H, Zang, J, Tan, X, Wang, Z, Du, M. | | Deposit date: | 2019-12-15 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crassostrea gigas ferritin mutant-D119G
To Be Published
|
|
8QRH
 
 | | Inactivated tick-borne encephalitis virus (TBEV) vaccine strain Sofjin-Chumakov | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, Small envelope protein M | | Authors: | Moiseenko, A.V, Zhang, Y, Vorovitch, M, Ivanova, A, Liu, Z, Osolodkin, D.I, Egorov, A, Ishmukhametov, A, Sokolova, O.S. | | Deposit date: | 2023-10-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structure of inactivated mature tick-borne encephalitis virus at 3.0 angstrom resolution.
Emerg Microbes Infect, 13, 2024
|
|
8RYT
 
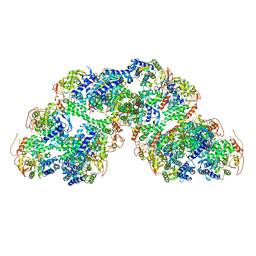 | | Structural characterization of Thogoto Virus nucleoprotein provides insights into RNA encapsidation and assembly | | Descriptor: | Nucleoprotein | | Authors: | Roske, Y, Mikirtumov, V, Daumke, O, Kudryashev, M, Dick, A. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural characterization of Thogoto Virus nucleoprotein provides insights into viral RNA encapsidation and RNP assembly.
Structure, 32, 2024
|
|
7R9J
 
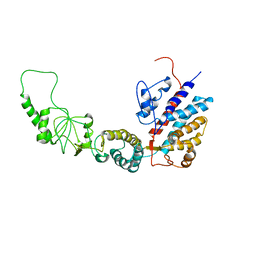 | | Methanococcus maripaludis chaperonin, open conformation 4 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
4R4Z
 
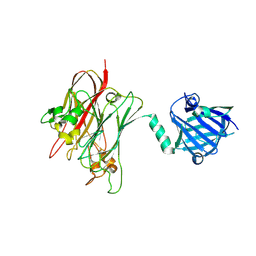 | | Structure of PNGF-II in P21 space group | | Descriptor: | PNGF-II | | Authors: | Sun, G, Yu, X, Celimuge, Wang, L, Li, M, Gan, J, Qu, D, Ma, J, Chen, L. | | Deposit date: | 2014-08-20 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Identification and
Characterization of a Novel Prokaryotic Peptide: N-glycosidase from
Elizabethkingia meningoseptica
J.Biol.Chem., 2015
|
|
8GVD
 
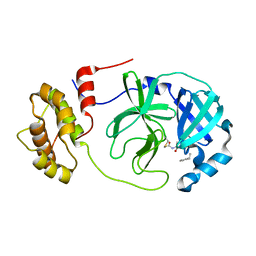 | | SARS-CoV-2 Mpro in complex with D-4-38 | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, 3C-like proteinase nsp5 | | Authors: | Liu, M, Huang, H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 Mpro in complex with D-4-38
To Be Published
|
|
7R9I
 
 | | Methanococcus maripaludis chaperonin, open conformation 2 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
4R69
 
 | | Lactate Dehydrogenase in complex with inhibitor compound 13 | | Descriptor: | (5R)-2-[(2-chlorophenyl)sulfanyl]-5-[2,6-dichloro-3-(tetrahydro-2H-pyran-4-ylamino)phenyl]-3-hydroxycyclohex-2-en-1-one, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Optimization of 5-(2,6-dichlorophenyl)-3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 25, 2014
|
|
7R9K
 
 | | Methanococcus maripaludis chaperonin, closed conformation 4 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
4RE2
 
 | | Different transition state conformations for the hydrolysis of beta-mannosides and beta-glucosides in the rice Os7BGlu26 family GH1 beta-mannosidase/beta-glucosidase | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tankrathok, A, Iglesias-Fernandez, J, Williams, R.J, Hakki, Z, Robinson, R.C, Hrmova, M, Rovira, C, Williams, S.J, Ketudat Cairns, J.R. | | Deposit date: | 2014-09-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Single Glycosidase Harnesses Different Pyranoside Ring Transition State Conformations for Hydrolysis of Mannosides and Glucosides
ACS CATALYSIS, 5, 2015
|
|
7R9H
 
 | | Methanococcus maripaludis chaperonin, open conformation 2 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9M
 
 | | Methanococcus maripaludis chaperonin, closed conformation 2 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
4RET
 
 | | Crystal structure of the Na,K-ATPase E2P-digoxin complex with bound magnesium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Gregersen, J.L, Laursen, M, Yatime, L, Nissen, P, Fedosova, N.U. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures and characterization of digoxin- and bufalin-bound Na+,K+-ATPase compared with the ouabain-bound complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6L0P
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Phosphoserine | | Descriptor: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
7R9E
 
 | | Methanococcus maripaludis chaperonin, open conformation 1 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7RAK
 
 | | Methanococcus maripaludis chaperonin complex in open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-07-01 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9O
 
 | | Methanococcus maripaludis chaperonin, closed conformation 1 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
6L22
 
 | | Crystal structure of CK2a1 H115Y with hematein | | Descriptor: | (6aR)-3,4,6a,10-tetrakis(oxidanyl)-6,7-dihydroindeno[2,1-c]chromen-9-one, Casein kinase II subunit alpha | | Authors: | Tsuyuguchi, M, Kinoshita, T. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12318087 Å) | | Cite: | Structural insights for producing CK2 alpha 1-specific inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4WEP
 
 | | Apo YehZ from Escerichia coli | | Descriptor: | Putative osmoprotectant uptake system substrate-binding protein OsmF | | Authors: | Kimber, M.S, Lang, S, Mendoza, K, Wood, J.M. | | Deposit date: | 2014-09-10 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | YehZYXW of Escherichia coli Is a Low-Affinity, Non-Osmoregulatory Betaine-Specific ABC Transporter.
Biochemistry, 54, 2015
|
|
8GKN
 
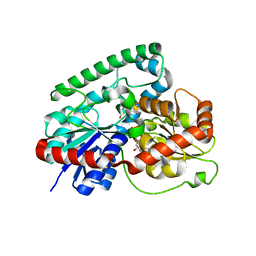 | | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with S-naringenin | | Descriptor: | NARINGENIN, UDP-glycosyltransferase 202A2, URIDINE-5'-DIPHOSPHATE | | Authors: | Arriaza, R.H, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with S-naringenin
To Be Published
|
|
8RWK
 
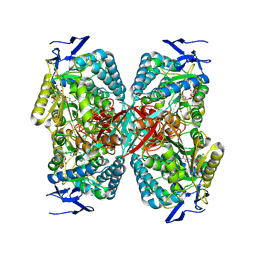 | | cryoEM structure of the central Ald4 filament determined by FilamentID | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium-activated aldehyde dehydrogenase, mitochondrial | | Authors: | Hugener, J, Xu, J, Wettstein, R, Ioannidi, L, Velikov, D, Wollweber, F, Henggeler, A, Matos, J, Pilhofer, M. | | Deposit date: | 2024-02-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | FilamentID reveals the composition and function of metabolic enzyme polymers during gametogenesis.
Cell, 187, 2024
|
|
7R9U
 
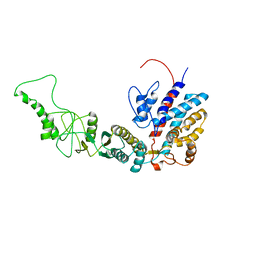 | | Methanococcus maripaludis chaperonin, closed conformation 3 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7RN0
 
 | |
