5UAM
 
 | | Structure of a new family of Polysaccharide lyase PL25-Ulvanlyase. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ulaganathan, T.S, Boniecki, M.T, Cygler, M. | | Deposit date: | 2016-12-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Ulvan-Degrading Polysaccharide Lyase Family: Structure and Catalytic Mechanism Suggests Convergent Evolution of Active Site Architecture.
ACS Chem. Biol., 12, 2017
|
|
5CXM
 
 | | Crystal structure of the cyanobacterial plasma membrane Rieske protein PetC3 from Synechocystis PCC 6803 | | Descriptor: | Cytochrome b6/f complex iron-sulfur subunit, FE2/S2 (INORGANIC) CLUSTER, NICKEL (II) ION, ... | | Authors: | Veit, S, Takeda, K, Miki, K, Roegner, M. | | Deposit date: | 2015-07-29 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterisation of the cyanobacterial PetC3 Rieske protein family.
Biochim. Biophys. Acta, 1857, 2016
|
|
6CFK
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with D-histidyl-CAM and bound to protein Y (YfiA) at 2.7A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
3UBW
 
 | | Complex of 14-3-3 isoform epsilon, a Mlf1 phosphopeptide and a small fragment hit from a FBDD screen | | Descriptor: | (3S)-pyrrolidin-3-ol, 14-3-3 protein epsilon, Myeloid leukemia factor 1, ... | | Authors: | Molzan, M, Weyand, M, Rose, R, Ottmann, C. | | Deposit date: | 2011-10-25 | | Release date: | 2012-01-25 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights of the MLF1/14-3-3 interaction.
Febs J., 279, 2012
|
|
7JIU
 
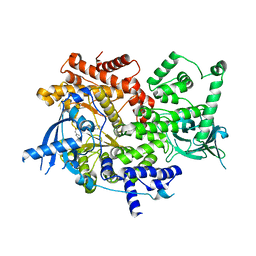 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 2F | | Descriptor: | (3S)-3-benzyl-5-[9-ethyl-8-(2-methylpyrimidin-5-yl)-9H-purin-6-yl]-3-methyl-1,3-dihydro-2H-indol-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Lesburg, C.A, Augustin, M. | | Deposit date: | 2020-07-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Optimization of Versatile Oxindoles as Selective PI3K delta Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
5UBP
 
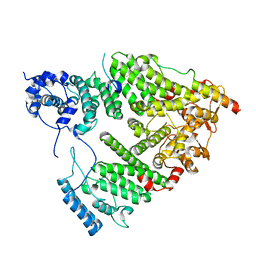 | | TREX2 M-region | | Descriptor: | 26S proteasome complex subunit SEM1, Leucine permease transcriptional regulator, Nuclear mRNA export protein THP1 | | Authors: | Stewart, M, Gordon, J. | | Deposit date: | 2016-12-21 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Sac3 RNA-binding M-region in the Saccharomyces cerevisiae TREX-2 complex.
Nucleic Acids Res., 45, 2017
|
|
5TPK
 
 | |
1JAQ
 
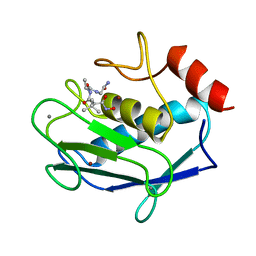 | | COMPLEX OF 1-HYDROXYLAMINE-2-ISOBUTYLMALONYL-ALA-GLY-NH2 WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (MET80 FORM), N-[(2R)-2-(hydroxycarbamoyl)-4-methylpentanoyl]-L-alanylglycinamide, ... | | Authors: | Grams, F, Reinemer, P, Powers, J.C, Kleine, T, Piper, M, Tschesche, H, Huber, R, Bode, W. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of human neutrophil collagenase complexed with peptide hydroxamate and peptide thiol inhibitors. Implications for substrate binding and rational drug design.
Eur.J.Biochem., 228, 1995
|
|
6C8H
 
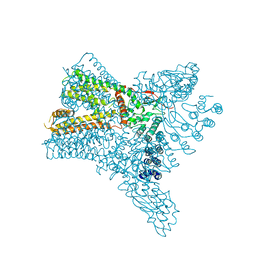 | | Crystal structure of Transient Receptor Potential (TRP) channel TRPV4 in the presence of gadolinium | | Descriptor: | GADOLINIUM ATOM, Transient receptor potential cation channel, subfamily V, ... | | Authors: | Deng, Z, Paknejad, N, Maksaev, G, Sala-Rabanal, M, Nichols, C.G, Hite, R.K, Yuan, P. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5OMZ
 
 | |
5D24
 
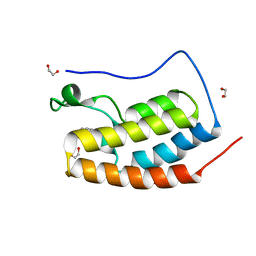 | | First bromodomain of BRD4 bound to inhibitor XD26 | | Descriptor: | 1,2-ETHANEDIOL, 4-acetyl-N-[3-(2-amino-2-oxoethoxy)phenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M, Gerhardt, S. | | Deposit date: | 2015-08-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5TSA
 
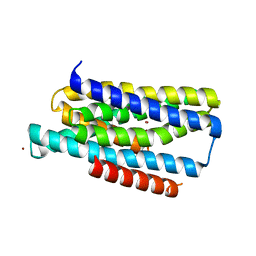 | | Crystal structure of the Zrt-/Irt-like protein from Bordetella bronchiseptica with bound Zn2+ | | Descriptor: | CADMIUM ION, Membrane protein, ZINC ION | | Authors: | Zhang, T, Fellner, M, Sui, D, Liu, J, Hu, J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway.
Sci Adv, 3, 2017
|
|
5TTH
 
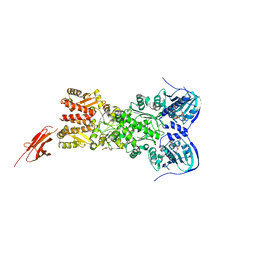 | | Heterodimeric SpyCatcher/SpyTag-fused zebrafish TRAP1 in ATP/ADP-hybrid state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, C-terminal SpyCatcher fusion of wildtype zebrafish TNF receptor-associated protein 1, ... | | Authors: | Elnatan, D, Betegon, M, Liu, Y, Agard, D.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-11-03 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Symmetry broken and rebroken during the ATP hydrolysis cycle of the mitochondrial Hsp90 TRAP1.
Elife, 6, 2017
|
|
5TSH
 
 | | PilB from Geobacter metallireducens bound to AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | McCallum, M, Tammam, S, Khan, A, Burrows, L, Howell, P.L. | | Deposit date: | 2016-10-28 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular mechanism of the type IVa pilus motors.
Nat Commun, 8, 2017
|
|
7NCB
 
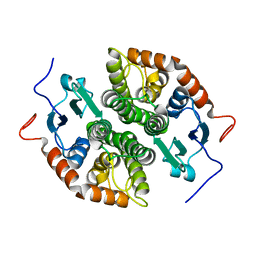 | | Glutathione-S-transferase GliG mutant H26A | | Descriptor: | Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC9
 
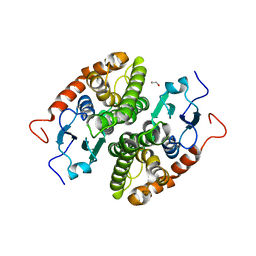 | | Glutathione-S-transferase GliG mutant H26N | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCM
 
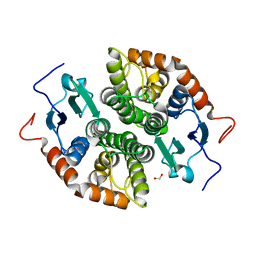 | | Glutathione-S-transferase GliG mutant E82A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCU
 
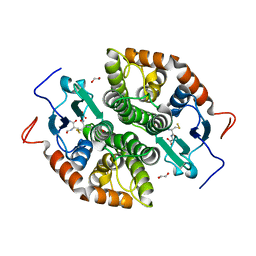 | |
5D3H
 
 | | First bromodomain of BRD4 bound to inhibitor XD29 | | Descriptor: | Bromodomain-containing protein 4, N-[5-(diethylsulfamoyl)-2-hydroxyphenyl]-3-ethyl-4-(hydroxyacetyl)-5-methyl-1H-pyrrole-2-carboxamide | | Authors: | Wohlwend, D, Huegle, M, Weitzel, G. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
7NCT
 
 | | Glutathione-S-transferase GliG mutant K127G | | Descriptor: | Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5TVX
 
 | |
7NC2
 
 | |
7NCE
 
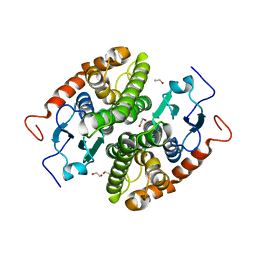 | | Glutathione-S-transferase GliG mutant N27A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6Y9R
 
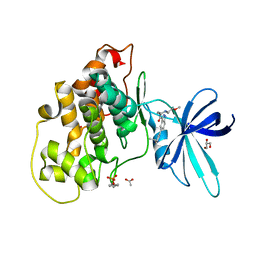 | | Crystal structure of GSK-3b in complex with the 1H-indazole-3-carboxamide inhibitor 2 | | Descriptor: | ACETATE ION, GLYCEROL, Glycogen synthase kinase-3 beta, ... | | Authors: | Krapp, S, Griessner, A, Blaesse, M, Buonfiglio, R, Ombrato, R. | | Deposit date: | 2020-03-10 | | Release date: | 2020-05-20 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Novel Imidazopyridine GSK-3 beta Inhibitors Supported by Computational Approaches.
Molecules, 25, 2020
|
|
7NCL
 
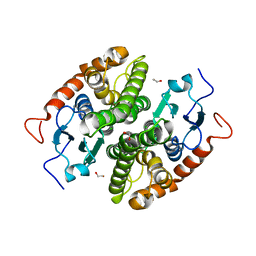 | | Glutathione-S-transferase GliG mutant E82Q | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
