7KKT
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84D mutant with pyruvate bound in the active site and R,R-bislysine bound at the allosteric site | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-28 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A TIGHT DIMER INTERFACE N84 RESIDUE, PLAYS A CRITICAL ROLE IN THE TRANSMISSION OF THE ALLOSTERIC INHIBITION SIGNALS IN Cj.DHDPS
To Be Published
|
|
6ETB
 
 | | Aerobic S262Y mutation of E. coli FLRD core | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Romao, C.V, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | A tyrosine mutation in E. coli flavodiiron protein increases radiation sensitivity in the crystal structure
To Be Published
|
|
6ETL
 
 | |
5T1O
 
 | | Solution-state NMR and SAXS structural ensemble of NPr (1-85) in complex with EIN-Ntr (170-424) | | Descriptor: | Phosphocarrier protein NPr, Phosphoenolpyruvate-protein phosphotransferase PtsP | | Authors: | Strickland, M, Stanley, A.M, Wang, G, Schwieters, C.D, Buchanan, S, Peterkofsky, A, Tjandra, N. | | Deposit date: | 2016-08-19 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure of the NPr:EIN(Ntr) Complex: Mechanism for Specificity in Paralogous Phosphotransferase Systems.
Structure, 24, 2016
|
|
5JMU
 
 | | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 | | Descriptor: | ACETATE ION, MAGNESIUM ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Tan, K, Gu, M, Clancy, S, Joachimiak, A. | | Deposit date: | 2016-04-29 | | Release date: | 2016-06-29 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 (CASP target)
To Be Published
|
|
6X0M
 
 | |
1FV3
 
 | | THE HC FRAGMENT OF TETANUS TOXIN COMPLEXED WITH AN ANALOGUE OF ITS GANGLIOSIDE RECEPTOR GT1B | | Descriptor: | ETHYL-TRIMETHYL-SILANE, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Fotinou, C, Emsley, P, Black, I, Ando, H, Ishida, H, Kiso, M, Sinha, K.A, Fairweather, N.F, Isaacs, N.W. | | Deposit date: | 2000-09-18 | | Release date: | 2001-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of tetanus toxin Hc fragment complexed with a synthetic GT1b analogue suggests cross-linking between ganglioside receptors and the toxin.
J.Biol.Chem., 276, 2001
|
|
6B4T
 
 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 4-methylpyridin-2-ol | | Descriptor: | 4-methylpyridin-2-ol, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 4-methylpyridin-2-ol
To Be Published
|
|
1PRR
 
 | | NMR-DERIVED THREE-DIMENSIONAL SOLUTION STRUCTURE OF PROTEIN S COMPLEXED WITH CALCIUM | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Bagby, S, Harvey, T.S, Eagle, S.G, Inouye, S, Ikura, M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR-derived three-dimensional solution structure of protein S complexed with calcium.
Structure, 2, 1994
|
|
5J8P
 
 | | Lys27-linked diubiquitin | | Descriptor: | D-ubiquitin, MAGNESIUM ION, ubiquitin | | Authors: | Pan, M, Gao, S, Zheng, Y. | | Deposit date: | 2016-04-08 | | Release date: | 2016-05-18 | | Last modified: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.554 Å) | | Cite: | Structure of Lys27-linked diubiquitin
To Be Published
|
|
6X1L
 
 | | The crystal structure of a functional uncharacterized protein KP1_0663 from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 | | Descriptor: | WbbZ protein | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-03 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
7KM0
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H59A mutant with pyruvate bound in the active site and R,R-bislysine bound at the allosteric site | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-11-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | H59 PLAYS THE MOST VITAL ROLE IN THE TRANSMISSION OF THE ALLOSTERIC INHIBITION SIGNALS IN Cj.DHDPS ENZYME
To Be Published
|
|
1PRA
 
 | |
1FTW
 
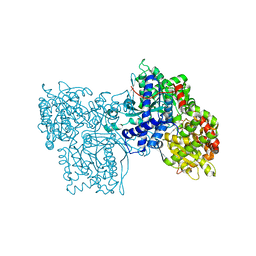 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | (5S,7R,8S,9S,10R)-3,8,9,10-tetrahydroxy-7-(hydroxymethyl)-6-oxa-1,3-diazaspiro[4.5]decane-2,4-dione, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-13 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
6WML
 
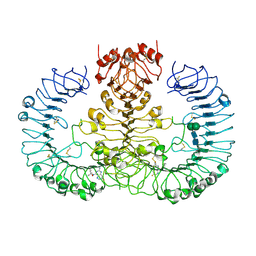 | | Human TLR8 bound to the potent agonist, GS-9688 (Selgantolimod) | | Descriptor: | (2R)-2-[(2-amino-7-fluoropyrido[3,2-d]pyrimidin-4-yl)amino]-2-methylhexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Appleby, T.C, Perry, J.K, Mish, M, Villasenor, A.G, Mackman, R.L. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of GS-9688 (Selgantolimod) as a Potent and Selective Oral Toll-Like Receptor 8 Agonist for the Treatment of Chronic Hepatitis B.
J.Med.Chem., 63, 2020
|
|
1FW9
 
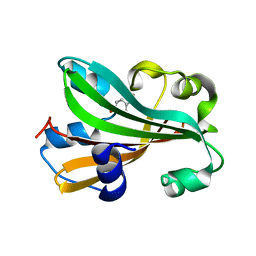 | | CHORISMATE LYASE WITH BOUND PRODUCT | | Descriptor: | CHORISMATE LYASE, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M, Vilker, V, Howard, A. | | Deposit date: | 2000-09-22 | | Release date: | 2001-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structure of chorismate lyase shows a new fold and a tightly retained product.
Proteins, 44, 2001
|
|
5T3M
 
 | |
7KM1
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H59N mutant with pyruvate bound in the active site and R,R-bislysine bound at the allosteric site | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-11-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | H59 PLAYS THE MOST VITAL ROLE IN THE TRANSMISSION OF THE ALLOSTERIC INHIBITION SIGNALS IN Cj.DHDPS ENZYME
To Be Published
|
|
6BAF
 
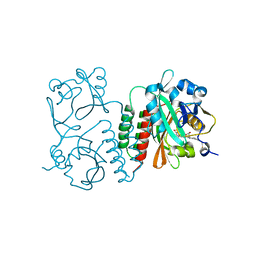 | | Structure of the chromophore binding domain of Stigmatella aurantiaca phytochrome P1, wild-type | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6F6H
 
 | |
6BAP
 
 | | Stigmatella aurantiaca bacterial phytochrome PAS-GAF-PHY, T289H mutant | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-14 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6EVS
 
 | | Characterization of 2-deoxyribosyltransferase from psychrotolerant bacterium Bacillus psychrosaccharolyticus: a suitable biocatalyst for the industrial synthesis of antiviral and antitumoral nucleosides | | Descriptor: | N-deoxyribosyltransferase | | Authors: | Fresco-Tabohada, A, Fernandez-Lucas, J, Acebal, C, Arroyo, M, Ramon, F, Mancheno, J.M, de la Mata, I. | | Deposit date: | 2017-11-02 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2'-Deoxyribosyltransferase from Bacillus psychrosaccharolyticus: A Mesophilic-Like Biocatalyst for the Synthesis of Modified Nucleosides from a Psychrotolerant Bacterium
Catalysts, 2019
|
|
6WNQ
 
 | | E. coli ATP Synthase State 2a | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
7KO1
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, E88D mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-11-06 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | THE ALLOSTERIC SITE RESIDUE, E88 INTERACTS WITH THE INHIBITORS TO TRANSMIT THE ALLOSTERIC INHIBITION SIGNALS IN Cj.DHDPS BY FORMING A HYDROGEN BOND.
To Be Published
|
|
6X5P
 
 | | Discovery of Hydroxy Pyrimidine Factor IXa Inhibitors | | Descriptor: | 3-chloro-4-{[5-hydroxy-6-(4-methylphenyl)pyrimidin-4-yl]amino}benzene-1-carboximidamide, CITRIC ACID, Coagulation factor IX, ... | | Authors: | Jayne, C.L, Andreani, T, Chan, T, Chelliah, M.V, Clasby, M.C, Dwyer, M, Eagen, K.A, Fried, S, Greenlee, W.J, Guo, Z, Hawes, B, Hruza, A, Ingram, R, Keertikar, K.M, Neelamkavil, S, Reichert, P, Xia, Y, Chackalamannil, S. | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Discovery of hydroxy pyrimidine Factor IXa inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
