5OMZ
 
 | |
6EO8
 
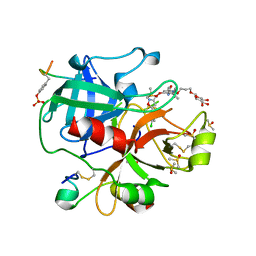 | | Crystal structure of thrombin in complex with a novel glucose-conjugated potent inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, Hirudin variant-2, N-(2-{[5-(5-chlorothiophen-2-yl)-1,2-oxazol-3-yl]methoxy}-6-[3-(beta-D-glucopyranosyloxy)propoxy]phenyl)-1-(propan-2-yl)piperidine-4-carboxamide, ... | | Authors: | Belviso, B.D, Caliandro, R, Aresta, B.M, De Candia, M, Altomare, C.D. | | Deposit date: | 2017-10-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | How a beta-D-glucoside side chain enhances binding affinity to thrombin of inhibitors bearing 2-chlorothiophene as P1 moiety: crystallography, fragment deconstruction study, and evaluation of antithrombotic properties.
J. Med. Chem., 57, 2014
|
|
5WE6
 
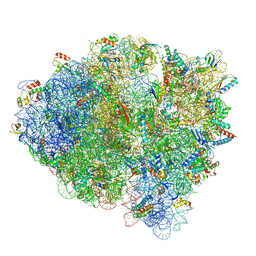 | | 70S ribosome-EF-Tu H84A complex with GTP and cognate tRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Frank, J. | | Deposit date: | 2017-07-07 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
6ESF
 
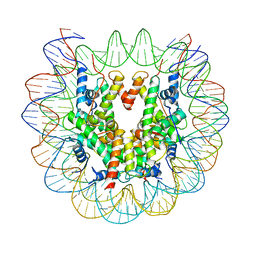 | | Nucleosome : Class 1 | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1QBN
 
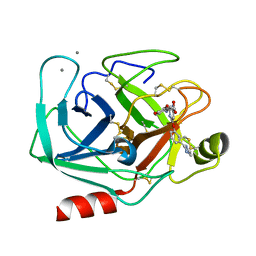 | |
2VKT
 
 | | HUMAN CTP SYNTHETASE 2 - GLUTAMINASE DOMAIN | | Descriptor: | CTP SYNTHASE 2 | | Authors: | Welin, M, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-28 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human Ctp Synthetase 2 - Glutaminase Domain
To be Published
|
|
5WL3
 
 | | Crystal structure of chalcone isomerase engineered from ancestral inference (ancR2) | | Descriptor: | CHLORIDE ION, Engineered Chalcone Isomerase ancR2 | | Authors: | Burke, J.R, Kaltenbach, M, Tawfik, D.S, Noel, J.P. | | Deposit date: | 2017-07-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution of chalcone isomerase from a noncatalytic ancestor.
Nat. Chem. Biol., 14, 2018
|
|
2A29
 
 | | The solution structure of the AMP-PNP bound nucleotide binding domain of KdpB | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Holo-form of the Nucleotide Binding Domain of the KdpFABC Complex from Escherichia coli Reveals a New Binding Mode
J.Biol.Chem., 281, 2006
|
|
1BWS
 
 | | CRYSTAL STRUCTURE OF GDP-4-KETO-6-DEOXY-D-MANNOSE EPIMERASE/REDUCTASE FROM ESCHERICHIA COLI A KEY ENZYME IN THE BIOSYNTHESIS OF GDP-L-FUCOSE | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (GDP-4-KETO-6-DEOXY-D-MANNOSE EPIMERASE/REDUCTASE) | | Authors: | Rizzi, M, Tonetti, M, Flora, A.D, Bolognesi, M. | | Deposit date: | 1998-09-25 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | GDP-4-keto-6-deoxy-D-mannose epimerase/reductase from Escherichia coli, a key enzyme in the biosynthesis of GDP-L-fucose, displays the structural characteristics of the RED protein homology superfamily.
Structure, 6, 1998
|
|
4LI5
 
 | | EGFR-K IN COMPLEX WITH N-[3-[[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]-4-methoxy-phenyl] Prop-2-enamide | | Descriptor: | Epidermal growth factor receptor, N-(3-{[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino}-4-methoxyphenyl)propanamide, SODIUM ION | | Authors: | Debreczeni, J.E, Seiffert, G.B, Kiefersauer, R, Augustin, M, Nagel, S, Ward, R, Anderton, M, Ashton, S, Bethel, P, Box, M, Butterworth, S, Colclough, N, Chroley, C, Chuaqui, C, Cross, D, Eberlein, C, Finlay, R, Hill, G, Grist, M, Klinowska, T, Lane, C, Martin, S, Orme, J, Smith, P, Wang, F, Waring, M. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure- and Reactivity-Based Development of Covalent Inhibitors of the Activating and Gatekeeper Mutant Forms of the Epidermal Growth Factor Receptor (EGFR).
J.Med.Chem., 56, 2013
|
|
5F91
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator (N-(5-(azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Fumarate hydratase class II, ... | | Authors: | Kasbekar, M, Fischer, G, Mott, B.T, Yasgar, A, Hyvonen, M, Boshoff, H.I, Abell, C, Barry, C.E, Thomas, C.J. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Selective small molecule inhibitor of the Mycobacterium tuberculosis fumarate hydratase reveals an allosteric regulatory site.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1HZU
 
 | | DOMAIN SWING UPON HIS TO ALA MUTATION IN NITRITE REDUCTASE OF PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRITE REDUCTASE | | Authors: | Brown, K, Cutruzzola, F, Brunori, M, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-01-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Domain swing upon His to Ala mutation in nitrite reductase of Pseudomonas aeruginosa.
J.Mol.Biol., 312, 2001
|
|
1HZV
 
 | | DOMAIN SWING UPON HIS TO ALA MUTATION IN NITRITE REDUCTASE OF PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRIC OXIDE, ... | | Authors: | Brown, K, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-01-26 | | Release date: | 2001-09-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Domain swing upon His to Ala mutation in nitrite reductase of Pseudomonas aeruginosa.
J.Mol.Biol., 312, 2001
|
|
5IOI
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
6EJN
 
 | |
4PPF
 
 | | Mycobacterium tuberculosis RecA citrate bound low temperature structure IIA-N | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
1QKK
 
 | | Crystal structure of the receiver domain and linker region of DctD from Sinorhizobium meliloti | | Descriptor: | C4-DICARBOXYLATE TRANSPORT TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Meyer, M.G, Park, S, Zeringue, L, Staley, M, Mckinstry, M, Kaufman, R.I, Zhang, H, Yan, D, Yennawar, N, Farber, G.K, Nixon, B.T. | | Deposit date: | 1999-07-23 | | Release date: | 2000-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A dimeric two-component receiver domain inhibits the sigma54-dependent ATPase in DctD.
Faseb J., 15, 2001
|
|
1J9M
 
 | | K38H mutant of Streptomyces K15 DD-transpeptidase | | Descriptor: | CHLORIDE ION, DD-transpeptidase, SODIUM ION | | Authors: | Fonze, E, Rhazi, N, Nguyen-Disteche, M, Charlier, P. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
5JDP
 
 | | E73V mutant of the human voltage-dependent anion channel | | Descriptor: | Voltage-dependent anion-selective channel protein 1 | | Authors: | Jaremko, M, Jaremko, L, Villinger, S, Schmidt, C, Giller, K, Griesinger, C, Becker, S, Zweckstetter, M. | | Deposit date: | 2016-04-17 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High-Resolution NMR Determination of the Dynamic Structure of Membrane Proteins.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6EE0
 
 | | Crystal Structure of SNX23 PX domain | | Descriptor: | Kinesin-like protein KIF16B | | Authors: | Chandra, M, Collins, B.M. | | Deposit date: | 2018-08-12 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities.
Nat Commun, 10, 2019
|
|
1ZOV
 
 | | Crystal Structure of Monomeric Sarcosine Oxidase from Bacillus sp. NS-129 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase | | Authors: | Nagata, K, Sasaki, H, Ohtsuka, J, Hua, M, Okai, M, Kubota, K, Kamo, M, Ito, K, Ichikawa, T, Koyama, Y, Tanokura, M. | | Deposit date: | 2005-05-14 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of monomeric sarcosine oxidase from Bacillus sp. NS-129 reveals multiple conformations at the active-site loop
PROC.JPN.ACAD.,SER.B, 81, 2005
|
|
4KNB
 
 | | C-Met in complex with OSI ligand | | Descriptor: | 7-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[3,2-c]pyridin-6-amine, GAMMA-BUTYROLACTONE, Hepatocyte growth factor receptor | | Authors: | Wang, J, Steinig, A.G, Li, A.H, Chen, X, Dong, H, Ferraro, C, Jin, M, Kadalbajoo, M, Kleinberg, A, Stolz, K.M, Tavares-Greco, P.A, Wang, T, Albertella, M.R, Peng, Y, Crew, L, Kahler, J. | | Deposit date: | 2013-05-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel 6-aminofuro[3,2-c]pyridines as potent, orally efficacious inhibitors of cMET and RON kinases.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4Q0R
 
 | | The catalytic core of Rad2 (complex I) | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*GP*TP*CP*AP*GP*AP*GP*CP*AP*AP*A)-3'), DNA repair protein RAD2 | | Authors: | Mietus, M, Nowak, E, Jaciuk, M, Kustosz, P, Nowotny, M. | | Deposit date: | 2014-04-02 | | Release date: | 2014-08-27 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the catalytic core of Rad2: insights into the mechanism of substrate binding.
Nucleic Acids Res., 42, 2014
|
|
4Q0Z
 
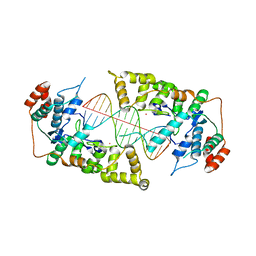 | | The catalytic core of Rad2 in complex with DNA substrate (complex III) | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*CP*TP*GP*AP*GP*AP*CP*AP*AP*GP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*CP*CP*TP*TP*GP*TP*CP*TP*CP*AP*GP*T)-3'), ... | | Authors: | Mietus, M, Nowak, E, Jaciuk, M, Kustosz, P, Nowotny, M. | | Deposit date: | 2014-04-02 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Crystal structure of the catalytic core of Rad2: insights into the mechanism of substrate binding.
Nucleic Acids Res., 42, 2014
|
|
6EM9
 
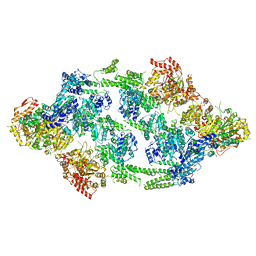 | | S.aureus ClpC resting state, asymmetric map | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
