1ASX
 
 | | APICAL DOMAIN OF THE CHAPERONIN FROM THERMOPLASMA ACIDOPHILUM | | Descriptor: | PHOSPHATE ION, THERMOSOME | | Authors: | Klumpp, M, Baumeister, W, Essen, L.-O. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin.
Cell(Cambridge,Mass.), 91, 1997
|
|
1P52
 
 | | Structure of Arginine kinase E314D mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, D-ARGININE, ... | | Authors: | Pruett, P.S, Azzi, A, Clark, S.A, Yousef, M.S, Gattis, J.L, Somasundarum, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase.
J.Biol.Chem., 278, 2003
|
|
1P50
 
 | | Transition state structure of an Arginine Kinase mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Pruett, P.S, Azzi, A, Clark, S.A, Yousef, M.S, Gattis, J.L, Somasundarum, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase.
J.Biol.Chem., 278, 2003
|
|
5TJK
 
 | | Crystal structure of GTA + A trisaccharide (native) | | Descriptor: | GLYCEROL, Histo-blood group ABO system transferase, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]octyl beta-D-galactopyranoside | | Authors: | Legg, M.S.G, Gagnon, S.M.L, Evans, S.V. | | Deposit date: | 2016-10-04 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structures and STD NMR mapping of human ABO(H) blood group glycosyltransferases in complex with trisaccharide reaction products suggest a molecular basis for product release.
Glycobiology, 27, 2017
|
|
5TJN
 
 | | Crystal structure of GTB + B trisaccharide (native) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Histo-blood group ABO system transferase, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]octyl beta-D-galactopyranoside | | Authors: | Legg, M.S.G, Gagnon, S.M.L, Evans, S.V. | | Deposit date: | 2016-10-04 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | High-resolution crystal structures and STD NMR mapping of human ABO(H) blood group glycosyltransferases in complex with trisaccharide reaction products suggest a molecular basis for product release.
Glycobiology, 27, 2017
|
|
4NG8
 
 | | Dialyzed HEW lysozyme batch crystallized in 1.9 M CsCl and collected at 100 K. | | Descriptor: | CESIUM ION, CHLORIDE ION, Lysozyme C | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NIX
 
 | | Crystal structure of trypsiligase (K60E/N143H/Y151H/D189K trypsin) orthorhombic form, zinc-bound | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Schoepfel, M, Parthier, C, Stubbs, M.T. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | N-terminal protein modification by substrate-activated reverse proteolysis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6XVR
 
 | | Human myelin protein P2 mutant L35S | | Descriptor: | Myelin P2 protein, PALMITIC ACID, SULFATE ION | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XXU
 
 | | Solution NMR structure of the native form of UbcH7 (UBE2L3) | | Descriptor: | Ubiquitin-conjugating enzyme E2 L3 | | Authors: | Marousis, K.D, Seliami, A, Birkou, M, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H,13C,15N backbone and side-chain resonance assignment of the native form of UbcH7 (UBE2L3) through solution NMR spectroscopy.
Biomol.Nmr Assign., 14, 2020
|
|
6XYK
 
 | |
6XCQ
 
 | | Erythromycin esterase EreC, mutant H289N in its closed conformation | | Descriptor: | EreC, S-1,2-PROPANEDIOL | | Authors: | Zielinski, M, Park, J, Berghuis, A.M. | | Deposit date: | 2020-06-09 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional insights into esterase-mediated macrolide resistance.
Nat Commun, 12, 2021
|
|
3AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, REFINED AVERAGE STRUCTURE | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-29 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
6XCS
 
 | |
6XHE
 
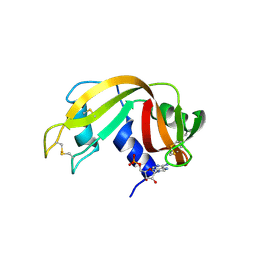 | | Structure of beta-prolinyl 5'-O-adenosine phosphoramidate | | Descriptor: | 5'-O-[(S)-[(3S)-3-carboxypyrrolidin-1-yl](hydroxy)phosphoryl]adenosine, ADENOSINE MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHF
 
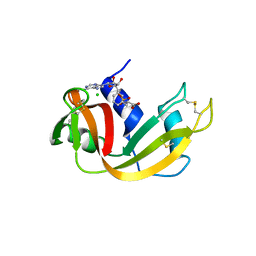 | | Structure of Phenylalanyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]amino]-3-phenyl-propanoic acid, CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHJ
 
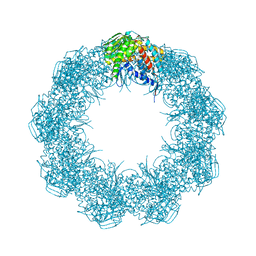 | | Cryo-EM structure of octadecameric TF55 (beta-only) complex from S. solfataricus bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Thermosome subunit beta | | Authors: | Zeng, Y.C, Sobti, M, Stewart, A.G. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural analysis of the Sulfolobus solfataricus TF55beta chaperonin by cryo-electron microscopy
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6XHI
 
 | |
6XA8
 
 | |
6XA6
 
 | |
6XHD
 
 | | Structure of Prolinyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-1-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]pyrrolidine-2-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XA7
 
 | |
6XOC
 
 | | Crystal structure of glVRC01 Fab in complex with anti-idiotypic iv4 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Aljedani, S.S, Weidle, C, Pancera, M. | | Deposit date: | 2020-07-06 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development of a VRC01-class germline targeting immunogen derived from anti-idiotypic antibodies.
Cell Rep, 35, 2021
|
|
4OTI
 
 | | Crystal Structure of PRK1 Catalytic Domain in Complex with Tofacitinib | | Descriptor: | 3-{(3R,4R)-4-methyl-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}-3-oxopropanenitrile, Serine/threonine-protein kinase N1 | | Authors: | Chamberlain, P.P, Delker, S, Pagarigan, B, Mahmoudi, A, Jackson, P, Abbassian, M, Muir, J, Raheja, N, Cathers, B. | | Deposit date: | 2014-02-13 | | Release date: | 2014-08-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structures of PRK1 in Complex with the Clinical Compounds Lestaurtinib and Tofacitinib Reveal Ligand Induced Conformational Changes.
Plos One, 9, 2014
|
|
7ZXY
 
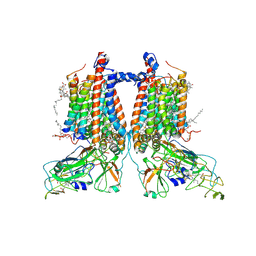 | | 3.15 Angstrom cryo-EM structure of the dimeric cytochrome b6f complex from Synechocystis sp. PCC 6803 with natively bound plastoquinone and lipid molecules. | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, CHLOROPHYLL A, Cytochrome B6, ... | | Authors: | Malone, L.A, Procter, M.S, Farmer, D.F, Swainsbury, D.J.K, Hawkings, F.R, Pastorelli, F, Emrich-Mills, T.Z, Siebert, A, Hunter, C.N, Hitchcock, A, Johnson, M.P. | | Deposit date: | 2022-05-23 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structures of the Synechocystis sp. PCC 6803 cytochrome b6f complex with and without the regulatory PetP subunit.
Biochem.J., 479, 2022
|
|
6XJJ
 
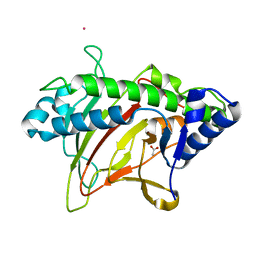 | | Structure of non-heme iron enzyme TropC: Radical tropolone biosynthesis | | Descriptor: | 2-oxoglutarate-dependent dioxygenase tropC, ACETATE ION, FE (III) ION, ... | | Authors: | Mallik, L, Doyon, T.J, Narayan, A.R.H, Koutmos, M. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Radical Tropolone Biosynthesis
Chemrxiv, 2020
|
|
