7KVC
 
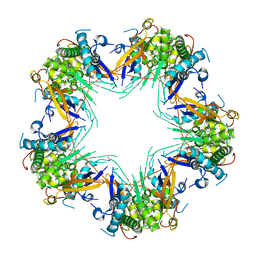 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (decamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
1R41
 
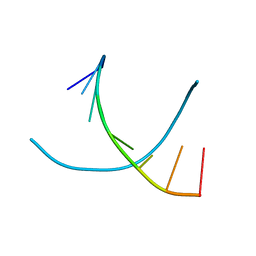 | | Crystal structures of d(Gm5CGm5CGCGC) and d(GCGCGm5CGm5C): Effects of methylation on alternating DNA octamers | | Descriptor: | 5'-D(*GP*CP*GP*CP*GP*(5CM)P*GP*(5CM))-3' | | Authors: | Shi, K, Pan, B, Tippin, D, Sundaralingam, M. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of d(Gm5)CGm5CGCGC) and d(GCGCGm5CGm5C): effects of methylation on alternating DNA octamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7KVD
 
 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (dodecamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
7L5O
 
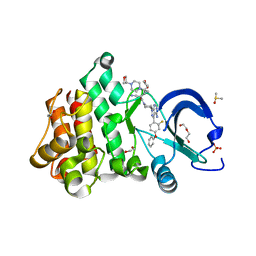 | | Crystal structure of the noncovalently bonded complex of rilzabrutinib with BTK | | Descriptor: | (2E)-2-{(3R)-3-[4-amino-3-(2-fluoro-4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidine-1-carbonyl}-4-methyl-4-[4-(oxetan-3-yl)piperazin-1-yl]pent-2-enenitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Bradshaw, J.M, Brameld, K.A, Mrosek, M, Lammens, A, Blaesse, M. | | Deposit date: | 2020-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The Discovery Rilzabrutinib (PRN1008): A Reversible Covalent BTK Inhibitor for Immune Mediated Diseases
To Be Published
|
|
7L5P
 
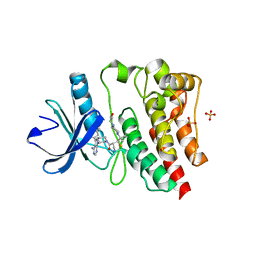 | | Crystal structure of the covalently bonded complex of rilzabrutinib with BTK | | Descriptor: | (2E)-2-{(3R)-3-[4-amino-3-(2-fluoro-4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidine-1-carbonyl}-4-methyl-4-[4-(oxetan-3-yl)piperazin-1-yl]pent-2-enenitrile, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Bradshaw, J.M, Brameld, K.A, Mrosek, M, Lammens, A, Blaesse, M. | | Deposit date: | 2020-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The Discovery Rilzabrutinib (PRN1008): A Reversible Covalent BTK Inhibitor for Immune Mediated Diseases
To Be Published
|
|
1R5W
 
 | | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T-cell activation | | Descriptor: | H-2 class II histocompatibility antigen, E-K alpha chain, MHC H2-IE-beta, ... | | Authors: | Krogsgaard, M, Prado, N, Adams, E, He, X, Chow, D.C, Wilson, D.B, Garcia, K.C, Davis, M.M. | | Deposit date: | 2003-10-13 | | Release date: | 2004-03-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation
Mol.Cell, 12, 2003
|
|
7KNV
 
 | | Solution NMR structure of CDHR3 extracellular domain EC1 | | Descriptor: | CALCIUM ION, Cadherin-related family member 3 | | Authors: | Lee, W, Tonelli, M, Frederick, R.O, Watters, K.E, Markley, J.L, Palmenberg, A.C. | | Deposit date: | 2020-11-06 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Determination of the CDHR3 Rhinovirus-C Binding Domain, EC1
Viruses, 13, 2021
|
|
1R4Q
 
 | | Shiga toxin | | Descriptor: | SHT cytotoxin A subunit, Shigella toxin chain B | | Authors: | Fraser, M.E, Fujinaga, M, Cherney, M.M, Melton-Celsa, A.R, Twiddy, E.M, O'Brien, A.D, James, M.N.G. | | Deposit date: | 2003-10-07 | | Release date: | 2004-05-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Shiga Toxin Type 2 (Stx2) from Escherichia coli O157:H7.
J.Biol.Chem., 279, 2004
|
|
4H18
 
 | | Three dimensional structure of corynomycoloyl tranferase C | | Descriptor: | Cmt1, MAGNESIUM ION | | Authors: | Huc, E, de Sousa D'Auria, C, Li de la Sierra-Gallay, I, Salmeron, C.H, van Tilbeurgh, H, Bayan, N, Houssin, C.H, Daffe, M, Tropis, M. | | Deposit date: | 2012-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Identification of a mycoloyl transferase selectively involved in o-acylation of polypeptides in corynebacteriales.
J.Bacteriol., 195, 2013
|
|
7L6K
 
 | | ApoL1 N-terminal domain | | Descriptor: | Apolipoprotein L1 | | Authors: | Holliday, M.J, Ultsch, M, Moran, P, Fairbrother, W.J, Kirchhofer, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
7LDO
 
 | | G150T Pseudomonas fluorescens isocyanide hydratase (G150T-3) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LG3
 
 | |
7L9S
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-2) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-04 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9W
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-3) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LA0
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-2) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LD6
 
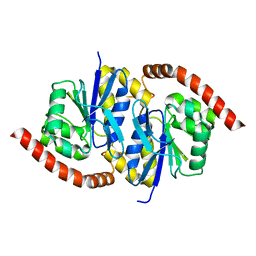 | | G150A Pseudomonas fluorescens isocyanide hydratase (G150A-1) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
4GY2
 
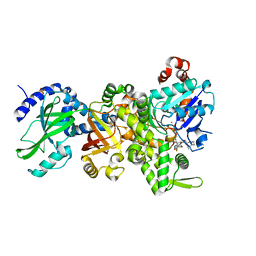 | | Crystal structure of apo-Ia-actin complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Tsurumura, T, Oda, M, Nagahama, M, Tsuge, H. | | Deposit date: | 2012-09-05 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1T7J
 
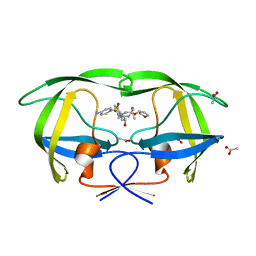 | | crystal structure of inhibitor amprenavir in complex with a multi-drug resistant variant of HIV-1 protease (L63P/V82T/I84V) | | Descriptor: | ACETATE ION, Pol Polyprotein, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | King, N.M, Prabu-Jeyabalan, M, Nalivaika, E.A, Wigerinck, P.B.T.P, De Bethune, M.-P, Schiffer, C.A. | | Deposit date: | 2004-05-10 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor
J.Med.Chem., 48, 2005
|
|
7L9Q
 
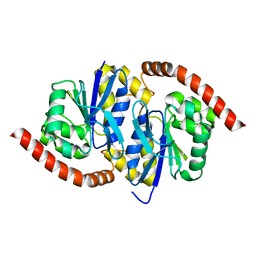 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-1) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-04 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LDQ
 
 | | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae R2846 | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-13 | | Release date: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae R2846
To Be Published
|
|
7L9Z
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-1) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LA3
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-3) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
1T8U
 
 | | Crystal Structure of human 3-O-Sulfotransferase-3 with bound PAP and tetrasaccharide substrate | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Moon, A.F, Edavettal, S.C, Krahn, J.M, Munoz, E.M, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1
J.Biol.Chem., 279, 2004
|
|
7LG0
 
 | |
1T8I
 
 | | Human DNA Topoisomerase I (70 Kda) In Complex With The Poison Camptothecin and Covalent Complex With A 22 Base Pair DNA Duplex | | Descriptor: | 4-ETHYL-4-HYDROXY-1,12-DIHYDRO-4H-2-OXA-6,12A-DIAZA-DIBENZO[B,H]FLUORENE-3,13-DIONE, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T)-3', ... | | Authors: | Staker, B.L, Feese, M.D, Cushman, M, Pommier, Y, Zembower, D, Stewart, L, Burgin, A.B. | | Deposit date: | 2004-05-12 | | Release date: | 2005-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex
J.Med.Chem., 48, 2005
|
|
