1V47
 
 | | Crystal structure of ATP sulfurylase from Thermus thermophillus HB8 in complex with APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, ATP sulfurylase, CHLORIDE ION, ... | | Authors: | Taguchi, Y, Sugishima, M, Fukuyama, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of a novel zinc-binding ATP sulfurylase from Thermus thermophilus HB8
Biochemistry, 43, 2004
|
|
6E85
 
 | | 1.25 Angstrom Resolution Crystal Structure of 4-hydroxythreonine-4-phosphate Dehydrogenase from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, D-threonate 4-phosphate dehydrogenase, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-27 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
1V5U
 
 | | Solution Structure of the C-terminal Pleckstrin Homology Domain of Sbf1 from Mouse | | Descriptor: | SET binding factor 1 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Pleckstrin Homology Domain of Sbf1 from Mouse
To be Published
|
|
6E8Q
 
 | | S. CEREVISIAE CYP51 COMPLEXED WITH Posaconazole | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Lanosterol 14-alpha demethylase, PENTAETHYLENE GLYCOL, ... | | Authors: | Tyndall, J.D, Keniya, M.V, Sabherwal, M, Monk, B.C. | | Deposit date: | 2018-07-30 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Azole Resistance Reduces Susceptibility to the Tetrazole Antifungal VT-1161.
Antimicrob. Agents Chemother., 63, 2019
|
|
1V65
 
 | | Solution structure of the Kruppel-associated box (KRAB) domain | | Descriptor: | RIKEN cDNA 2610044O15 | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Kruppel-associated box (KRAB) domain
To be Published
|
|
1V7Z
 
 | | creatininase-product complex | | Descriptor: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-12-26 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
7RR5
 
 | | Structure of ribosomal complex bound with Rbg1/Tma46 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Zeng, F, Li, X, Pires-Alves, M, Chen, X, Hawk, C.W, Jin, H. | | Deposit date: | 2021-08-09 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells.
Cell Rep, 37, 2021
|
|
1V7F
 
 | | Solution structure of phrixotoxin 1 | | Descriptor: | Phrixotoxin 1 | | Authors: | Chagot, B, Escoubas, P, Villegas, E, Bernard, C, Ferrat, G, Corzo, G, Lazdunski, M, Darbon, H. | | Deposit date: | 2003-12-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Phrixotoxin 1, a specific peptide inhibitor of Kv4 potassium channels from the venom of the theraphosid spider Phrixotrichus auratus
Protein Sci., 13, 2004
|
|
1V9V
 
 | | Solution structure of putative domain of human KIAA0561 protein | | Descriptor: | KIAA0561 protein | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-03 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of putative domain of human KIAA0561 protein
To be Published
|
|
7RED
 
 | | Holo Hemophilin from A. baumannii | | Descriptor: | Hemophilin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bateman, T.J, Shah, M, Moraes, T.F. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A Slam-dependent hemophore contributes to heme acquisition in the bacterial pathogen Acinetobacter baumannii.
Nat Commun, 12, 2021
|
|
1V84
 
 | | Crystal structure of human GlcAT-P in complex with N-acetyllactosamine, Udp, and Mn2+ | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID, MANGANESE (II) ION, ... | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
1V8E
 
 | |
6ECJ
 
 | |
7RE4
 
 | | Apo Hemophilin from A. baumannii | | Descriptor: | Hemophilin | | Authors: | Bateman, T.J, Shah, M, Moraes, T.F. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A Slam-dependent hemophore contributes to heme acquisition in the bacterial pathogen Acinetobacter baumannii.
Nat Commun, 12, 2021
|
|
7REA
 
 | | Apo Hemophilin from A. baumannii | | Descriptor: | Hemophilin | | Authors: | Bateman, T.J, Shah, M, Moraes, T.F. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A Slam-dependent hemophore contributes to heme acquisition in the bacterial pathogen Acinetobacter baumannii.
Nat Commun, 12, 2021
|
|
1UMK
 
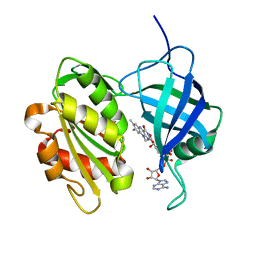 | | The Structure of Human Erythrocyte NADH-cytochrome b5 Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase | | Authors: | Bando, S, Takano, T, Yubisui, T, Shirabe, K, Takeshita, M, Horii, C, Nakagawa, A. | | Deposit date: | 2003-10-03 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human erythrocyte NADH-cytochrome b5 reductase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ULY
 
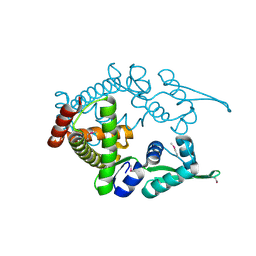 | | Crystal structure analysis of the ArsR homologue DNA-binding protein from P. horikoshii OT3 | | Descriptor: | hypothetical protein PH1932 | | Authors: | Itou, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-09-17 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the PH1932 protein, a unique archaeal ArsR type winged-HTH transcription factor from Pyrococcus horikoshii OT3
Proteins, 70, 2008
|
|
1UNF
 
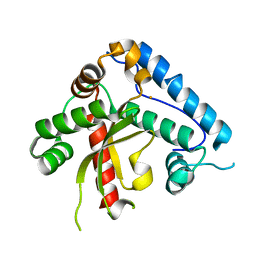 | | The crystal structure of the eukaryotic FeSOD from Vigna unguiculata suggests a new enzymatic mechanism | | Descriptor: | FE (III) ION, IRON SUPEROXIDE DISMUTASE | | Authors: | Munoz, I.G, Moran, J.F, Becana, M, Montoya, G. | | Deposit date: | 2003-09-10 | | Release date: | 2004-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Crystal Structure of an Eukaryotic Iron Superoxide Dismutase Suggests Intersubunit Cooperation During Catalysis
Protein Sci., 14, 2005
|
|
6EHL
 
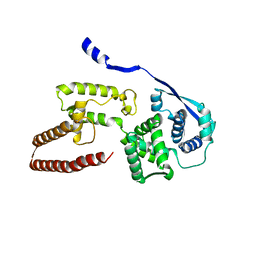 | | Model of the Ebola virus nucleoprotein in recombinant nucleocapsid-like assemblies | | Descriptor: | Nucleoprotein | | Authors: | Wan, W, Kolesnikova, L, Clarke, M, Koehler, A, Noda, T, Becker, S, Briggs, J.A.G. | | Deposit date: | 2017-09-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure and assembly of the Ebola virus nucleocapsid.
Nature, 551, 2017
|
|
1URV
 
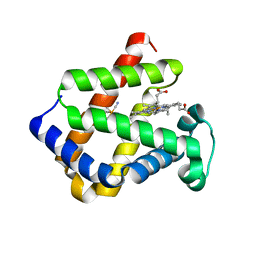 | | Crystal structure of cytoglobin: the fourth globin type discovered in man displays heme hexa-coordination | | Descriptor: | CYTOGLOBIN, HEXACYANOFERRATE(3-), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | de Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2003-11-11 | | Release date: | 2004-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Cytoglobin: The Fourth Globin Type Discovered in Man Displays Heme Hexa-Coordination.
J.Mol.Biol., 336, 2004
|
|
7RPV
 
 | |
1UYO
 
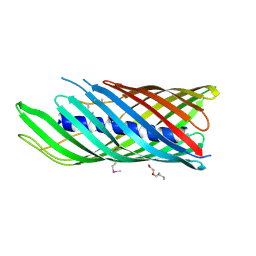 | | Translocator domain of autotransporter NalP from Neisseria meningitidis | | Descriptor: | NALP, PENTAETHYLENE GLYCOL MONODECYL ETHER | | Authors: | Oomen, C.J, Van Ulsen, P, Van Gelder, P, Feijen, M, Tommassen, J, Gros, P. | | Deposit date: | 2004-03-02 | | Release date: | 2004-03-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Translocator Domain of a Bacterial Autotransporter
Embo J., 23, 2004
|
|
1V1F
 
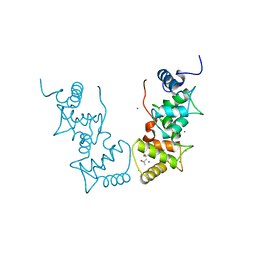 | | Structure of the Arabidopsis thaliana SOS3 complexed with Calcium(II) and Manganese(II) ions | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCINEURIN B-LIKE PROTEIN 4, CALCIUM ION, ... | | Authors: | Sanchez-Barrena, M.J, Martinez-Ripoll, M, Zhu, J.K, Albert, A. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of the Arabidopsis Thaliana SOS3: Molecular Mechanism of Sensing Calcium for Salt Stress Response
J.Mol.Biol., 345, 2005
|
|
4WEX
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation Y168S | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
6E6R
 
 | | 1.50 A resolution structure of the C-terminally truncated [2Fe-2S] ferredoxin (Bfd) R26E mutant from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin-associated ferredoxin, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Lovell, S, Wijerathne, H, Battaile, K.P, Yao, H, Wang, Y, Rivera, M. | | Deposit date: | 2018-07-25 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bfd, a New Class of [2Fe-2S] Protein That Functions in Bacterial Iron Homeostasis, Requires a Structural Anion Binding Site.
Biochemistry, 57, 2018
|
|
