9FQ9
 
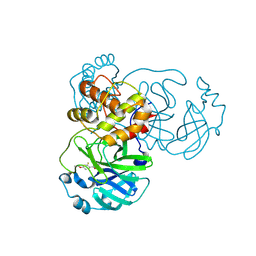 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalently bound inhibitor PSB-21110 (compound 29b in publication) | | Descriptor: | (5-chloranylpyridin-3-yl) 4-ethoxy-2-fluoranyl-benzoate, BROMIDE ION, Non-structural protein 11 | | Authors: | Strater, N, Claff, T, Sylvester, K, Oneto, A, Guetschow, M, Mueller, C.E. | | Deposit date: | 2024-06-19 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Nonpeptidic Irreversible Inhibitors of SARS-CoV-2 Main Protease with Potent Antiviral Activity.
J.Med.Chem., 67, 2024
|
|
9FTH
 
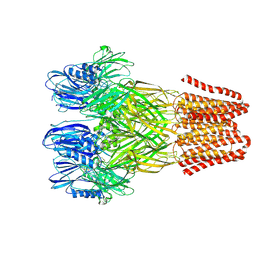 | | Open conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10mM Ca2+ | | Descriptor: | Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
9FY4
 
 | | Crystal structure of Heme-Oxygenase mutant I143K from Corynebacterium diphtheriae complexed with Cobalt-porphyrine (HumO-Co(III)) | | Descriptor: | CHLORIDE ION, COBALT (II) ION, PROTOPORPHYRIN IX CONTAINING CO, ... | | Authors: | Labidi, R.J, Faivre, B, Carpentier, P, Perard, J, Gotico, P, Li, Y, Atta, M, Fontecave, M. | | Deposit date: | 2024-07-02 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.839 Å) | | Cite: | Light-Activated Artificial CO 2 -Reductase: Structure and Activity.
J.Am.Chem.Soc., 2024
|
|
6ZJY
 
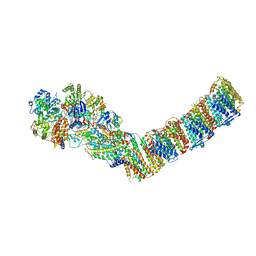 | | Respiratory complex I from Thermus thermophilus, NAD+ dataset, minor state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, NADH-quinone oxidoreductase subunit 1, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
9GD3
 
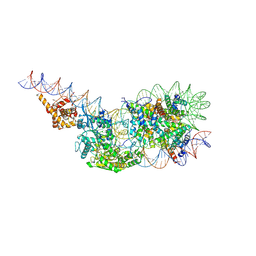 | | Structure of a mononucleosome bound by one copy of Chd1 with the DBD on the exit-side DNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
6ZI8
 
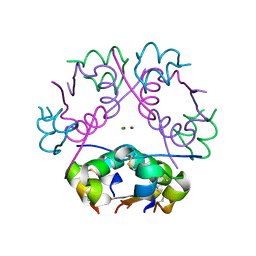 | | X-ray diffraction structure of bovine insulin at 2.3 A resolution | | Descriptor: | CHLORIDE ION, Insulin, ZINC ION | | Authors: | Housset, D, Ling, W.L, Bacia-Verloop, M, Zander, U, McCarthy, A.A, Schoehn, G. | | Deposit date: | 2020-06-25 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
9GG0
 
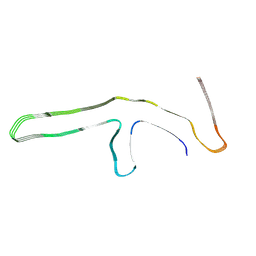 | | P301L tau filaments from human brain | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2024-08-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
8CZ6
 
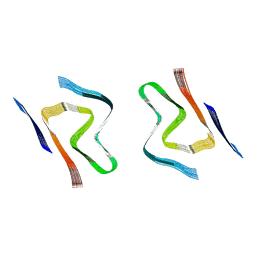 | | CryoEM structure of amplified alpha-synuclein fibril class A type I with extended core from DLB case X | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
9JHV
 
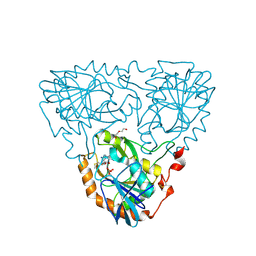 | | Crystal Structure of 5'-Deoxy-5'-methylthioadenosine phosphorylase from Aeropyrum pernix complex with 5'-Deoxy-5'-methylthioadenosine 343K | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Iizuka, Y, Kikuchi, M, Yamauchi, T, Tsunoda, M. | | Deposit date: | 2024-09-10 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of 5'-Deoxy-5'-methylthioadenosine phosphorylase from Aeropyrum pernix complex with 5'-Deoxy-5'-methylthioadenosine 353K
To Be Published
|
|
9GLE
 
 | | Jumonji domain-containing protein 2A with crystallization epitope mutations A91T:T93S | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-08-27 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To be published
|
|
6EK4
 
 | | PaxB from Photorhabdus luminescens | | Descriptor: | PaxB, SODIUM ION | | Authors: | Braeuning, B, Groll, M. | | Deposit date: | 2017-09-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of the two-component alpha-helical pore-forming toxin YaxAB.
Nat Commun, 9, 2018
|
|
9GRE
 
 | |
6EL1
 
 | | YaxAB pore complex | | Descriptor: | YaxA, YaxB | | Authors: | Braeuning, B, Bertosin, E, Dietz, H, Groll, M. | | Deposit date: | 2017-09-27 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure and mechanism of the two-component alpha-helical pore-forming toxin YaxAB.
Nat Commun, 9, 2018
|
|
6ENT
 
 | | Structure of the rat RKIP variant delta143-146 | | Descriptor: | CHLORIDE ION, Phosphatidylethanolamine-binding protein 1, ZINC ION | | Authors: | Koelmel, W, Hirschbeck, M, Schindelin, H, Lorenz, K, Kisker, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
9FTG
 
 | | Closed conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10 mM Ca2+ | | Descriptor: | CALCIUM ION, Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
9FTI
 
 | | Open conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10mM EDTA | | Descriptor: | Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
9C61
 
 | | Crystal structure of the human LRRK2 WDR domain in complex with CACHE1193-26 | | Descriptor: | (4R)-4-[4-(5-fluoro-1H-indol-3-yl)piperidine-1-carbonyl]piperidin-2-one, Non-specific serine/threonine protein kinase, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Kutera, M, Ilyassov, O, Seitova, A, Loppnau, P, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Halabelian, L. | | Deposit date: | 2024-06-07 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the human LRRK2 WDR domain in complex with CACHE1193-26
To be published
|
|
8CYX
 
 | | CryoEM structure of amplified alpha-synuclein fibril class A type I with compact core from DLB case III | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
9EPP
 
 | | Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Giq heterotrimer | | Descriptor: | 11,20-Ethanoretinal, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tejero, O, Pamula, F, Koyanagi, M, Nagata, T, Afanasyev, P, Das, I, Deupi, X, Sheves, M, Terakita, A, Schertler, G.F.X, Rodrigues, M.J, Tsai, C.-J. | | Deposit date: | 2024-03-19 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Active state structures of a bistable visual opsin bound to G proteins.
Nat Commun, 15, 2024
|
|
6E4D
 
 | | Atomic structure of Mycobacterium tuberculosis DppA | | Descriptor: | Periplasmic dipeptide-binding lipoprotein DPPA, VAL-VAL-VAL-ALA | | Authors: | Ko, Y, Mitra, A, Niederweis, M, Cingolani, G. | | Deposit date: | 2018-07-17 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.252 Å) | | Cite: | Heme and hemoglobin utilization by Mycobacterium tuberculosis.
Nat Commun, 10, 2019
|
|
9FQA
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalently bound inhibitor PSB-21101 (compound 30b in publication) | | Descriptor: | (5-chloranylpyridin-3-yl) 2-fluoranyl-4-phenylmethoxy-benzoate, BROMIDE ION, MAGNESIUM ION, ... | | Authors: | Strater, N, Claff, T, Sylvester, K, Oneto, A, Guetschow, M, Mueller, C.E. | | Deposit date: | 2024-06-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Nonpeptidic Irreversible Inhibitors of SARS-CoV-2 Main Protease with Potent Antiviral Activity.
J.Med.Chem., 67, 2024
|
|
9EZ8
 
 | | Cryo-EM structure of the icosahedral lumazine synthase from Vicia faba. | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Chee, M, Trapani, S, Hoh, F, Lai Kee Him, J, Yvon, M, Blanc, S, Bron, P. | | Deposit date: | 2024-04-11 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of the icosahedral lumazine synthase from Vicia faba.
To Be Published
|
|
9FS7
 
 | | Coxsackievirus A16 3C protease in C2 2 21 spacegroup | | Descriptor: | Genome polyprotein | | Authors: | Fairhead, M, Lithgo, R, MacLean, M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Coxsackievirus A16 3C protease in C2 2 21 spacegroup
To Be Published
|
|
6E4L
 
 | | The structure of the N-terminal domain of human clathrin heavy chain 1 (nTD) in complex with ES9 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(4-nitrophenyl)thiophene-2-sulfonamide, ACETATE ION, ... | | Authors: | Dejonghe, W, Sharma, I, Denoo, B, Munck, S.D, Bulut, H, Mylle, E, Vasileva, M, Lu, Q, Savatin, D.V, Mishev, K, Nerinckx, W, Staes, A, Drozdzecki, A, Audenaert, D, Madder, A, Friml, J, Damme, D.V, Gevaert, K, Haucke, V, Savvides, S, Winne, J, Russinova, E. | | Deposit date: | 2018-07-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Disruption of endocytosis through chemical inhibition of clathrin heavy chain function.
Nat.Chem.Biol., 15, 2019
|
|
9EPQ
 
 | | Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Giq heterotrimer | | Descriptor: | 11,20-Ethanoretinal, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tejero, O, Pamula, F, Koyanagi, M, Nagata, T, Afanasyev, P, Das, I, Deupi, X, Sheves, M, Terakita, A, Schertler, G.F.X, Rodrigues, M.J, Tsai, C.-J. | | Deposit date: | 2024-03-19 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Active state structures of a bistable visual opsin bound to G proteins.
Nat Commun, 15, 2024
|
|
