7O5X
 
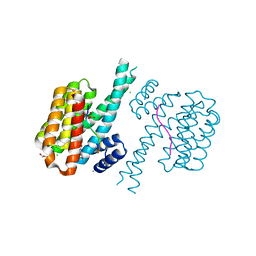 | |
7NZK
 
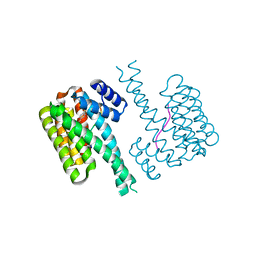 | |
6M6S
 
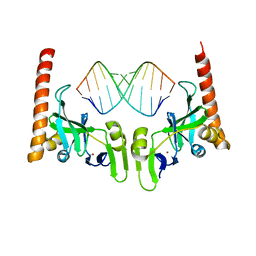 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) C-terminal domain with 5'-ppp 12-mer dsRNA | | 分子名称: | Dicer Related Helicase, RNA (5'-R(*(GTP)P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ZINC ION | | 著者 | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | 登録日 | 2020-03-16 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
7O6I
 
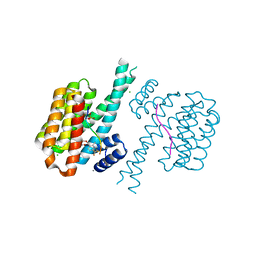 | |
1BHU
 
 | | THE 3D STRUCTURE OF THE STREPTOMYCES METALLOPROTEINASE INHIBITOR, SMPI, ISOLATED FROM STREPTOMYCES NIGRESCENS TK-23, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | METALLOPROTEINASE INHIBITOR | | 著者 | Tate, S, Ohno, A, Seeram, S.S, Hiraga, K, Oda, K, Kainosho, M. | | 登録日 | 1998-06-10 | | 公開日 | 1999-01-06 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the Streptomyces metalloproteinase inhibitor, SMPI, isolated from Streptomyces nigrescens TK-23: another example of an ancestral beta gamma-crystallin precursor structure.
J.Mol.Biol., 282, 1998
|
|
7O3Q
 
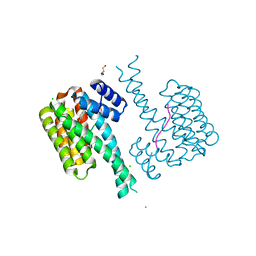 | |
7O6O
 
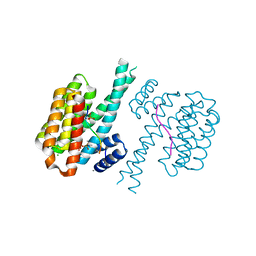 | |
1TOV
 
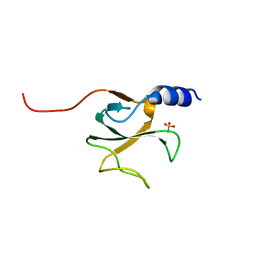 | | Structural genomics of Caenorhabditis elegans: CAP-GLY domain of F53F4.3 | | 分子名称: | Hypothetical protein F53F4.3 in chromosome V, SULFATE ION | | 著者 | Li, S, Finley, J, Liu, Z.J, Qiu, S.H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, Delucas, L.J, Richardson, D, Richardson, J, Wang, B.C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2004-06-15 | | 公開日 | 2004-07-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal Structure of the Cytoskeleton-Associated Protein Glycine-Rich (CAP-Gly) Domain
J.Biol.Chem., 277, 2002
|
|
1X3C
 
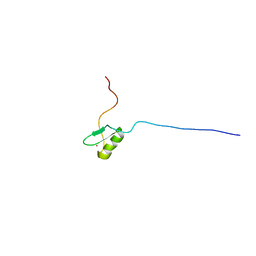 | | Solution structure of the C2H2 type zinc-binding domain of human zinc finger protein 292 | | 分子名称: | ZINC ION, Zinc finger protein 292 | | 著者 | Yoneyama, M, Tomizawa, T, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-05-02 | | 公開日 | 2005-11-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the C2H2 type zinc-binding domain of human zinc finger protein 292
To be Published
|
|
7O5A
 
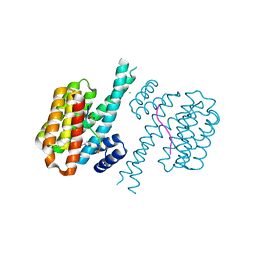 | |
7O6G
 
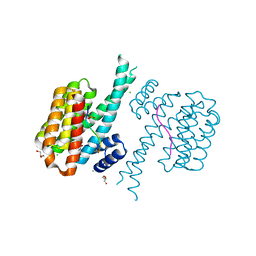 | |
1X47
 
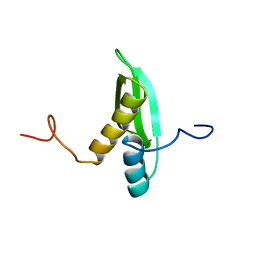 | | Solution structure of DSRM domain in DGCR8 protein | | 分子名称: | DGCR8 protein | | 著者 | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-05-14 | | 公開日 | 2005-11-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of DSRM domain in DGCR8 protein
To be Published
|
|
1X4P
 
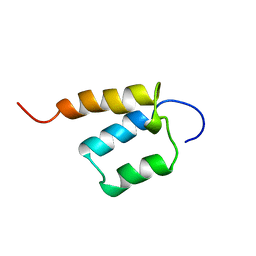 | | Solution structure of SURP domain in SFRS14 protei | | 分子名称: | Putative splicing factor, arginine/serine-rich 14 | | 著者 | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-05-14 | | 公開日 | 2005-11-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of SURP domain in SFRS14 protei
To be Published
|
|
7OH5
 
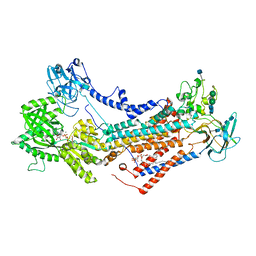 | | Cryo-EM structure of Drs2p-Cdc50p in the E1-AlFx-ADP state | | 分子名称: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Timcenko, M, Dieudonne, T, Montigny, C, Boesen, T, Lyons, J.A, Lenoir, G, Nissen, P. | | 登録日 | 2021-05-09 | | 公開日 | 2021-06-09 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of substrate-independent phosphorylation in a P4-ATPase lipid flippase
J.Mol.Biol., 2021
|
|
1X5Z
 
 | | Solution structure of the fibronectin type-III domain of human protein tyrosine phosphatase, receptor type, D isoform 4 variant | | 分子名称: | Receptor-type tyrosine-protein phosphatase delta | | 著者 | Yoneyama, M, Saito, K, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-05-17 | | 公開日 | 2005-11-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the fibronectin type-III domain of human protein tyrosine phosphatase, receptor type, D isoform 4 variant
To be Published
|
|
5AON
 
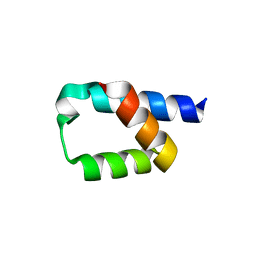 | | Crystal structure of the conserved N-terminal domain of Pex14 from Trypanosoma brucei | | 分子名称: | PEROXIN 14, SULFATE ION | | 著者 | Obita, T, Sugawara, Y, Mizuguchi, M, Watanabe, Y, Kawaguchi, K, Imanaka, T. | | 登録日 | 2015-09-11 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.646 Å) | | 主引用文献 | Characterization of the Interaction between Trypanosoma Brucei Pex5P and its Receptor Pex14P.
FEBS Lett., 590, 2016
|
|
1WQM
 
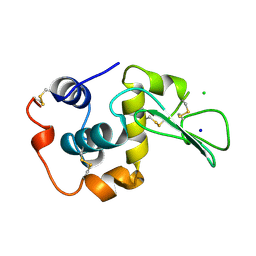 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, SODIUM ION | | 著者 | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1998-02-09 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
7NRK
 
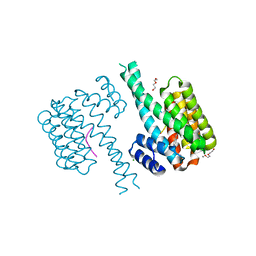 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1002F1 | | 分子名称: | 14-3-3 protein sigma, 4-(4-methylimidazol-1-yl)benzaldehyde, CALCIUM ION, ... | | 著者 | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | 登録日 | 2021-03-04 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7NRL
 
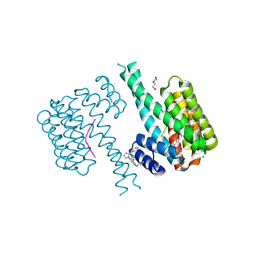 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1032 | | 分子名称: | 14-3-3 protein sigma, 2-(hydroxymethyl)-5-(2-phenylimidazol-1-yl)phenol, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | 登録日 | 2021-03-04 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
6PBE
 
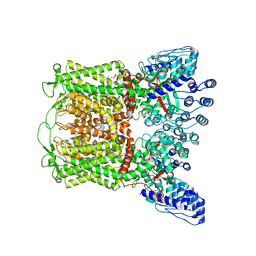 | | ZINC17988990-bound TRPV5 in nanodiscs | | 分子名称: | (4-oxo-5-phenyl-3,4-dihydrothieno[2,3-d]pyrimidin-2-yl)methyl 3-(3-oxo-2,3-dihydro-4H-1,4-benzoxazin-4-yl)propanoate, Transient receptor potential cation channel subfamily V member 5 | | 著者 | Hughes, T.E.T, Rosario, J.S.D, Kapoor, A, Yazici, A.T, Fluck, E.C, Filizola, M, Rohacs, T, Moiseenkova-Bell, V.Y. | | 登録日 | 2019-06-13 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Structure-based characterization of novel TRPV5 inhibitors.
Elife, 8, 2019
|
|
1WQO
 
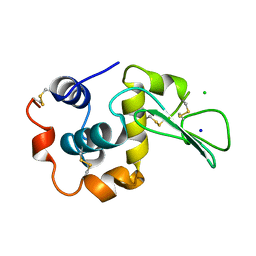 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, SODIUM ION | | 著者 | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1998-02-09 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
7NSV
 
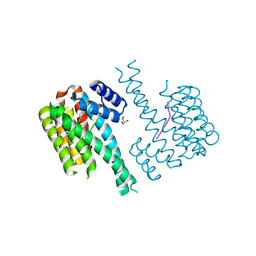 | | 14-3-3 sigma with p65 (RelA) binding site pS45 and covalently bound PC2046 | | 分子名称: | 1-(3-bromanyl-4-methyl-phenyl)-2-(2-bromophenyl)imidazole, 14-3-3 protein sigma, GLYCEROL, ... | | 著者 | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | 登録日 | 2021-03-08 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
1WQR
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, SODIUM ION | | 著者 | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1998-02-09 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
7NTK
 
 | |
1WQP
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, SODIUM ION | | 著者 | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1998-02-09 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
