7PFX
 
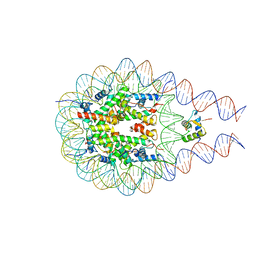 | |
7PEY
 
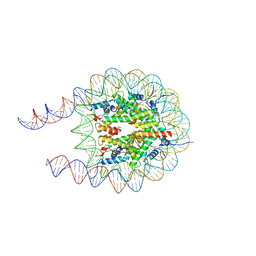 | |
7PFC
 
 | |
1AXH
 
 | | ATRACOTOXIN-HVI FROM HADRONYCHE VERSUTA (AUSTRALIAN FUNNEL-WEB SPIDER, NMR, 20 STRUCTURES | | 分子名称: | ATRACOTOXIN-HVI | | 著者 | Fletcher, J.I, O'Donoghue, S.I, Nilges, M, King, G.F. | | 登録日 | 1996-11-04 | | 公開日 | 1997-11-12 | | 最終更新日 | 2021-02-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of a novel insecticidal neurotoxin, omega-atracotoxin-HV1, from the venom of an Australian funnel web spider.
Nat.Struct.Biol., 4, 1997
|
|
7PEX
 
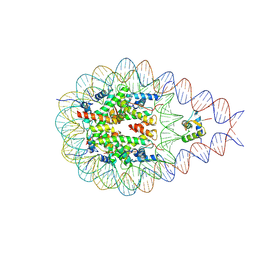 | |
5BMO
 
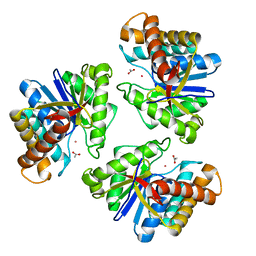 | | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus | | 分子名称: | ACETATE ION, POTASSIUM ION, Putative uncharacterized protein LnmX | | 著者 | Osipiuk, J, Hatzos-Skintges, C, Cuff, M, Endres, M, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-05-22 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus.
to be published
|
|
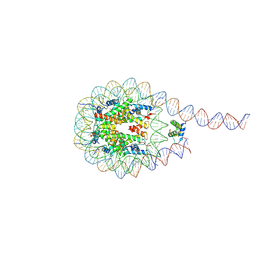
7PEZ
 
 | |
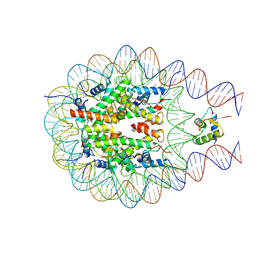
7PF3
 
 | |
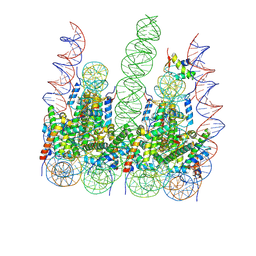
7PET
 
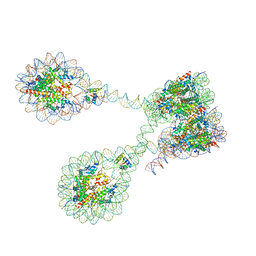 | | The 4x177 nucleosome array containing H1 | | 分子名称: | DNA (702-MER), Histone H1.4, Histone H2A type 1-B/E, ... | | 著者 | Dombrowski, M, Cramer, P. | | 登録日 | 2021-08-11 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Histone H1 binding to nucleosome arrays depends on linker DNA length and trajectory.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PF5
 
 | |
7PF4
 
 | |
7PFU
 
 | |
7PF6
 
 | |
7PEV
 
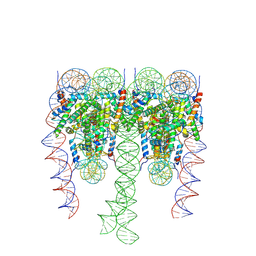 | |
5AX1
 
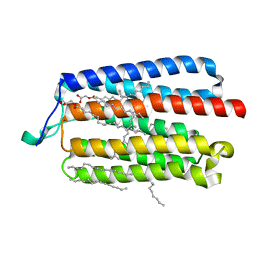 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.80 angstrom | | 分子名称: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | 著者 | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | 登録日 | 2015-07-10 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7PF2
 
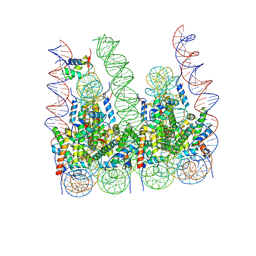 | |
7PFA
 
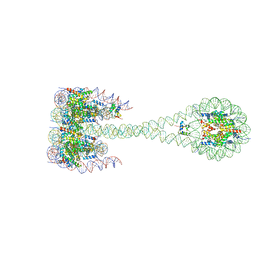 | |
7PFF
 
 | |
7PEU
 
 | |
7PFD
 
 | |
7PFT
 
 | |
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | 著者 | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | 登録日 | 2013-07-29 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.556 Å) | | 主引用文献 | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6AZC
 
 | |
7PFE
 
 | |
1BAH
 
 | | A TWO DISULFIDE DERIVATIVE OF CHARYBDOTOXIN WITH DISULFIDE 13-33 REPLACED BY TWO ALPHA-AMINOBUTYRIC ACIDS, NMR, 30 STRUCTURES | | 分子名称: | CHARYBDOTOXIN | | 著者 | Song, J, Gilquin, B, Jamin, N, Guenneugues, M, Dauplais, M, Vita, C, Menez, A. | | 登録日 | 1996-06-06 | | 公開日 | 1997-01-11 | | 最終更新日 | 2020-01-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of a two-disulfide derivative of charybdotoxin: structural evidence for conservation of scorpion toxin alpha/beta motif and its hydrophobic side chain packing.
Biochemistry, 36, 1997
|
|
