8VXE
 
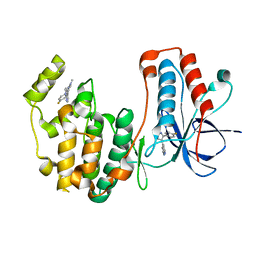 | | Structure of p38 alpha (Mitogen-activated protein kinase 14) complexed with inhibitor 6 | | 分子名称: | (4M)-4-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-1H-pyrrolo[2,3-b]pyridine, Mitogen-activated protein kinase 14 | | 著者 | Blaesse, M, Steinbacher, S, Shaffer, P.L, Sharma, S, Thompson, A.A. | | 登録日 | 2024-02-04 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
8VZM
 
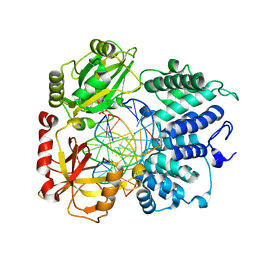 | | DNA Ligase 1 captured with pre-step 3 ligation at the rA:T nicksite | | 分子名称: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | 著者 | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | 登録日 | 2024-02-11 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
6YI7
 
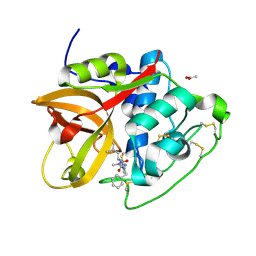 | | Structure of cathepsin B1 from Schistosoma mansoni (SmCB1) in complex with an azanitrile inhibitor | | 分子名称: | 1-[(2~{S})-1-[[iminomethyl(methyl)amino]-methyl-amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-3-(phenylmethyl)urea, ACETATE ION, Cathepsin B-like peptidase (C01 family) | | 著者 | Jilkova, A, Rezacova, P, Pachl, P, Fanfrlik, J, Rubesova, P, Guetschow, M, Mares, M. | | 登録日 | 2020-04-01 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Azanitrile Inhibitors of the SmCB1 Protease Target Are Lethal to Schistosoma mansoni : Structural and Mechanistic Insights into Chemotype Reactivity.
Acs Infect Dis., 7, 2021
|
|
5NNY
 
 | |
6Y91
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with NADH | | 分子名称: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | 登録日 | 2020-03-06 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity.
Commun Biol, 4, 2021
|
|
8VQ2
 
 | | HSV1 polymerase ternary complex with dsDNA and compound 44 | | 分子名称: | 2-(4-bromophenyl)-N-(3-methoxy-4-{[(4S)-2-oxo-1,3-oxazolidin-4-yl]methyl}phenyl)acetamide, DNA (5'-D(P*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*AP*GP*GP*AP*T)-3'), ... | | 著者 | Hayes, R.P, Heo, M.R, Plotkin, M. | | 登録日 | 2024-01-17 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (3.829 Å) | | 主引用文献 | Discovery of Broad-Spectrum Herpes Antiviral Oxazolidinone Amide Derivatives and Their Structure-Activity Relationships.
Acs Med.Chem.Lett., 15, 2024
|
|
8VU5
 
 | |
7OMM
 
 | | Cryo-EM structure of N. gonorhoeae LptDE in complex with ProMacrobodies (MBPs have not been built de novo) | | 分子名称: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ProMacrobody 21,Maltodextrin-binding protein, ... | | 著者 | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | 登録日 | 2021-05-24 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
3QSM
 
 | | Crystal structure for the MSOX.chloride binary complex | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase | | 著者 | Kommoju, P, Chen, Z, Bruckner, R.C, Mathews, F.S, Jorns, M.S. | | 登録日 | 2011-02-21 | | 公開日 | 2011-06-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Probing oxygen activation sites in two flavoprotein oxidases using chloride as an oxygen surrogate.
Biochemistry, 50, 2011
|
|
4UND
 
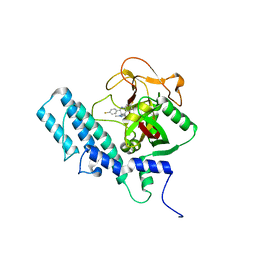 | | HUMAN ARTD1 (PARP1) - CATALYTIC DOMAIN IN COMPLEX WITH INHIBITOR TALAZOPARIB | | 分子名称: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, POLY [ADP-RIBOSE] POLYMERASE 1, SODIUM ION | | 著者 | Karlberg, T, Thorsell, A.G, Ekblad, T, Klepsch, M, Pinto, A.F, Tresaugues, L, Moche, M, Schuler, H. | | 登録日 | 2014-05-27 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
1JRR
 
 | | HUMAN PLASMINOGEN ACTIVATOR INHIBITOR-2.[LOOP (66-98) DELETIONMUTANT] COMPLEXED WITH PEPTIDE MIMIckING THE REACTIVE CENTER LOOP | | 分子名称: | BETA-MERCAPTOETHANOL, PLASMINOGEN ACTIVATOR INHIBITOR-2 | | 著者 | Jankova, L, Harrop, S.J, Saunders, D.N, Andrews, J.L, Bertram, K.C, Gould, A.R, Baker, M.S, Curmi, P.M.G. | | 登録日 | 2001-08-15 | | 公開日 | 2001-09-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF THE COMPLEX OF PLASMINOGEN ACTIVATOR INHIBITOR 2 WITH A PEPTIDE MIMICKING THE REACTIVE CENTER LOOP
J.Biol.Chem., 276, 2001
|
|
8X34
 
 | |
8XCR
 
 | |
8XCV
 
 | |
8XCW
 
 | |
8XD0
 
 | |
8XD1
 
 | |
8XD5
 
 | |
8XCQ
 
 | |
8XCU
 
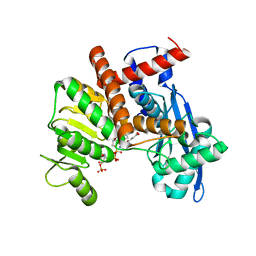 | | Cryo-EM structure of Glutamate dehydrogenase from Thermococcus profundus incorporating NADPH, AKG and GLU in the steady stage of reaction | | 分子名称: | 2-OXOGLUTARIC ACID, GAMMA-L-GLUTAMIC ACID, Glutamate dehydrogenase, ... | | 著者 | Wakabayashi, T, Oide, M, Nakasako, M. | | 登録日 | 2023-12-10 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | CryoEM-sampling of metastable conformations appearing in cofactor-ligand association and catalysis of glutamate dehydrogenase.
Sci Rep, 14, 2024
|
|
7OIG
 
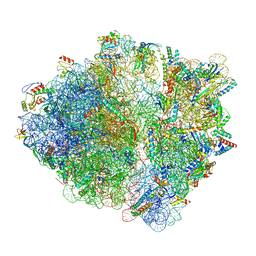 | | CspA-27 cotranslational folding intermediate 3 | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | 登録日 | 2021-05-11 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
8XCX
 
 | |
8XD4
 
 | |
8XD2
 
 | |
8XCO
 
 | |
