3DBJ
 
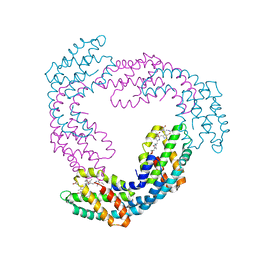 | | Allophycocyanin from Thermosynechococcus vulcanus | | 分子名称: | Allophycocyanin, PHYCOCYANOBILIN | | 著者 | Adir, N, Klartag, M, McGregor, A, David, L. | | 登録日 | 2008-06-01 | | 公開日 | 2008-11-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Allophycocyanin Trimer Stability and Functionality Are Primarily Due to Polar Enhanced Hydrophobicity of the Phycocyanobilin Binding Pocket
J.Mol.Biol., 384, 2008
|
|
5S36
 
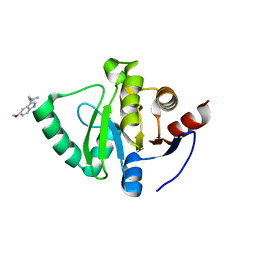 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z2856434938 | | 分子名称: | 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.058 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3PMZ
 
 | | Crystal Structure of the Complex of Acetylcholine Binding Protein and d-tubocurarine | | 分子名称: | (1beta,1'alpha)-7',12'-dihydroxy-6,6'-dimethoxy-2,2',2'-trimethyltubocuraran-2'-ium, MAGNESIUM ION, Soluble acetylcholine receptor | | 著者 | Talley, T.T, Harel, M, Yamauchi, J.G, Radic, Z, Hansen, S, Huxford, T, Taylor, P.W. | | 登録日 | 2010-11-18 | | 公開日 | 2011-10-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | The Curare Alkaloids: Analyzing the Poses of Complexes with the Acetylcholine Binding Protein in Relation to Structure and Binding Energies
To be Published
|
|
5S2W
 
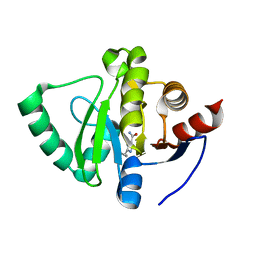 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z1407672867 | | 分子名称: | 2-cyclopropyl-1~{H}-imidazole-4-carboxamide, CHLORIDE ION, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.081 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3DC7
 
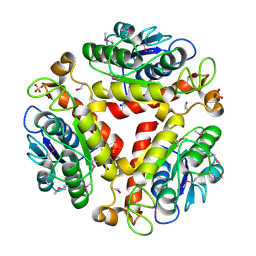 | | Crystal structure of the protein Q88SR8 from Lactobacillus plantarum. Northeast Structural Genomics consortium target LpR109. | | 分子名称: | MAGNESIUM ION, Putative uncharacterized protein lp_3323, SODIUM ION, ... | | 著者 | Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Seetharaman, J, Zhao, L, Mao, L, Ciccosanti, C, Xiao, R, Nair, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-06-03 | | 公開日 | 2008-08-05 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Crystal structure of the protein Q88SR8 from Lactobacillus plantarum. Northeast Structural Genomics consortium target LpR109. (CASP Target)
To be Published
|
|
5S3P
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z1238477790 | | 分子名称: | N-(cyclopentanecarbonyl)-L-alanine, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
1L3G
 
 | | NMR Structure of the DNA-binding Domain of Cell Cycle Protein, Mbp1(2-124) from Saccharomyces cerevisiae | | 分子名称: | TRANSCRIPTION FACTOR Mbp1 | | 著者 | Nair, M, McIntosh, P.B, Frenkiel, T.A, Kelly, G, Taylor, I.A, Smerdon, S.J, Lane, A.N. | | 登録日 | 2002-02-27 | | 公開日 | 2003-02-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of the DNA-Binding Domain of the Cell Cycle Protein Mbp1 from Saccharomyces cerevisiae
Biochemistry, 42, 2003
|
|
5S3C
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z2856434937 | | 分子名称: | (4-acetylphenoxy)acetic acid, DIMETHYL SULFOXIDE, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.185 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3PO7
 
 | | Human monoamine oxidase B in complex with zonisamide | | 分子名称: | 1-(1,2-benzoxazol-3-yl)methanesulfonamide, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Binda, C, Aldeco, M, Mattevi, A, Edmondson, D.E. | | 登録日 | 2010-11-22 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Interactions of monoamine oxidases with the antiepileptic drug zonisamide: specificity of inhibition and structure of the human monoamine oxidase B complex
J.Med.Chem., 54, 2011
|
|
5S41
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with NCL-00023825 | | 分子名称: | 4-bromanyl-1~{H}-pyridin-2-one, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.186 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2P9D
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | 分子名称: | FORMIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Yamamoto, H, Taketa, M, Morikawa, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-25 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
4L66
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina with highly ordered water structure in the substrate binding site | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, rRNA N-glycosidase | | 著者 | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2013-06-12 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of Ribosome inactivating protein from Momordica balsamina with highly ordered water structure in the substrate binding site
To be Published
|
|
5S3T
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with POB0128 | | 分子名称: | (1R,2S)-2-(thiophen-3-yl)cyclopentane-1-carboxylic acid, DIMETHYL SULFOXIDE, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.085 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3FVD
 
 | | Crystal structure of a member of enolase superfamily from ROSEOVARIUS NUBINHIBENS ISM complexed with magnesium | | 分子名称: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | 著者 | Malashkevich, V.N, Rutter, M, Bain, K.T, Lau, C, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-01-15 | | 公開日 | 2009-01-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of a member of enolase superfamily from ROSEOVARIUS NUBINHIBENS ISM complexed with magnesium
to be published
|
|
3GC1
 
 | | Crystal structure of bovine lactoperoxidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | 著者 | Singh, A.K, Singh, N, Sinha, M, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | 登録日 | 2009-02-21 | | 公開日 | 2009-03-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mode of binding of the tuberculosis prodrug isoniazid to heme peroxidases: binding studies and crystal structure of bovine lactoperoxidase with isoniazid at 2.7 A resolution.
J.Biol.Chem., 285, 2010
|
|
5S4A
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z955123498 | | 分子名称: | 4-AMINOPYRIDINE, DIMETHYL SULFOXIDE, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.081 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4Q6Q
 
 | | Structural analysis of the Zn-form II of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | 分子名称: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | 著者 | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | 登録日 | 2014-04-23 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2PB6
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | 分子名称: | FORMIC ACID, GLYCEROL, Probable diphthine synthase, ... | | 著者 | Yamamoto, H, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-28 | | 公開日 | 2007-10-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
3PEO
 
 | | Crystal structure of acetylcholine binding protein complexed with metocurine | | 分子名称: | 6,6',7',12'-tetramethoxy-2,2,2',2'-tetramethyltubocuraran-2,2'-diium, Soluble acetylcholine receptor | | 著者 | Talley, T.T, Harel, M, Yamauchi, G.J, Radic, Z, Hansen, S, Huxford, T, Taylor, P.W. | | 登録日 | 2010-10-27 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The curare alkaloids: analyzing the poses of complexes with the acetylcholine binding protein in relation to structure and binding energetics
To be Published
|
|
4L6U
 
 | | Crystal structure of AF1868: Cmr1 subunit of the Cmr RNA silencing complex | | 分子名称: | Putative uncharacterized protein | | 著者 | Sun, J, Jeon, J.H, Shin, M, Shin, H.C, Oh, B.H, Kim, J.S. | | 登録日 | 2013-06-12 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and CRISPR RNA-binding site of the Cmr1 subunit of the Cmr interference complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3GCL
 
 | | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid | | 分子名称: | 2-(ACETYLOXY)BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Singh, A.K, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2009-02-22 | | 公開日 | 2009-03-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Binding modes of aromatic ligands to mammalian heme peroxidases with associated functional implications: crystal structures of lactoperoxidase complexes with acetylsalicylic acid, salicylhydroxamic acid, and benzylhydroxamic acid
J.Biol.Chem., 284, 2009
|
|
4L7B
 
 | | Structure of keap1 kelch domain with (1S,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid | | 分子名称: | (1S,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | 著者 | Jnoff, E, Brookfield, F, Albrecht, C, Barker, J.J, Barker, O, Beaumont, E, Bromidge, S, Brooks, M, Ceska, T, Courade, J.P, Crabbe, T, Duclos, S, Fryatt, T, Jigorel, E, Kwong, J, Sands, Z, Smith, M.A. | | 登録日 | 2013-06-13 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
4Q4A
 
 | | Improved model of AMP-PNP bound TM287/288 | | 分子名称: | ABC transporter, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Hohl, M, Gruetter, M.G, Seeger, M.A. | | 登録日 | 2014-04-14 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5S1I
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with EN300-301084 | | 分子名称: | 5-amino-2-methyl-1,3-oxazole-4-carbonitrile, DIMETHYL SULFOXIDE, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.068 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
1L7G
 
 | | Crystal structure of E119G mutant influenza virus neuraminidase in complex with BCX-1812 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | 著者 | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | 登録日 | 2002-03-15 | | 公開日 | 2002-05-29 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
