4KAA
 
 | | Crystal structure of the halotag2 protein at the resolution 2.3A, Northeast Structural Genomics Consortium (NESG) target OR150 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Haloalkane dehalogenase, SODIUM ION | | 著者 | Kuzin, A, Lew, S, Neklesa, T.K, Noblin, D, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Crews, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-04-22 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.276 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target OR150
To be Published
|
|
5RE4
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1129283193 | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-methylpyridin-3-yl)acetamide | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4PSW
 
 | | Crystal structure of histone acetyltransferase complex | | 分子名称: | COENZYME A, Histone H4 type VIII, Histone acetyltransferase type B catalytic subunit, ... | | 著者 | Yang, M, Li, Y. | | 登録日 | 2014-03-08 | | 公開日 | 2014-07-09 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
5REL
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102340 | | 分子名称: | 1-{4-[(3-methylphenyl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2021-02-24 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
3U1N
 
 | | Structure of the catalytic core of human SAMHD1 | | 分子名称: | PHOSPHATE ION, SAM domain and HD domain-containing protein 1, ZINC ION | | 著者 | Goldstone, D.C, Ennis-Adeniran, V, Walker, P.A, Haire, L.F, Webb, M, Taylor, I.A. | | 登録日 | 2011-09-30 | | 公開日 | 2011-11-16 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase
Nature, 480, 2011
|
|
3DFN
 
 | |
5RF1
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with NCL-00023830 | | 分子名称: | 3C-like proteinase, 4-bromobenzene-1-sulfonamide, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFF
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102704 | | 分子名称: | 1-{4-[(4-chlorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2021-02-24 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4PUL
 
 | | tRNA-Guanine Transglycosylase (TGT) Mutant D102N in Complex with 6-Amino-2-(methylamino)-1H,7H,8H-imidazo[4,5-g]quinazolin-8-one | | 分子名称: | 6-amino-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | 著者 | Neeb, M, Heine, A, Klebe, G. | | 登録日 | 2014-03-13 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.654 Å) | | 主引用文献 | Chasing Protons: How Isothermal Titration Calorimetry, Mutagenesis, and pKa Calculations Trace the Locus of Charge in Ligand Binding to a tRNA-Binding Enzyme.
J.Med.Chem., 57, 2014
|
|
4KBD
 
 | | DNA STRUCTURE OF A MUTATED KB SITE | | 分子名称: | DNA (5'-D(*CP*CP*TP*GP*GP*AP*AP*AP*GP*TP*GP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*CP*TP*CP*AP*CP*TP*TP*TP*CP*CP*AP*GP*G)-3') | | 著者 | Tisne, C, Hartmann, B, Delepierre, M. | | 登録日 | 1998-11-30 | | 公開日 | 1999-10-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NF-kappa B binding mechanism: a nuclear magnetic resonance and modeling study of a GGG --> CTC mutation.
Biochemistry, 38, 1999
|
|
5RFV
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102306 | | 分子名称: | 1-[4-(thiophene-2-carbonyl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2021-02-24 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
6HYX
 
 | | THE GLIC PENTAMERIC LIGAND-GATED ION CHANNEL MUTANT Y197F-P250C | | 分子名称: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Proton-gated ion channel, ... | | 著者 | Hu, H.D, Delarue, M. | | 登録日 | 2018-10-22 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3PEH
 
 | | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of a thienopyrimidine derivative | | 分子名称: | 2-amino-4-{2,4-dichloro-5-[2-(diethylamino)ethoxy]phenyl}-N-ethylthieno[2,3-d]pyrimidine-6-carboxamide, Endoplasmin homolog, SULFATE ION | | 著者 | Wernimont, A.K, Tempel, W, Hutchinson, A, Weadge, J, Cossar, D, MacKenzie, F, Vedadi, M, Senisterra, G, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P.G, Fairlamb, A.H, MacKenzie, C, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | 登録日 | 2010-10-26 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of a thienopyrimidine derivative
To be Published
|
|
3PIL
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in reduced form | | 分子名称: | ACETATE ION, Peptide methionine sulfoxide reductase | | 著者 | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | 登録日 | 2010-11-07 | | 公開日 | 2011-02-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
4PMR
 
 | | Crystal structure of the Mycobacterium tuberculosis Tat-secreted protein Rv2525c in complex with HEPES (monoclinic crystal form II) | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SODIUM ION, Tat-secreted protein Rv2525c | | 著者 | Bellinzoni, M, Haouz, A, Shepard, W, Alzari, P.M. | | 登録日 | 2014-05-22 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural studies suggest a peptidoglycan hydrolase function for the Mycobacterium tuberculosis Tat-secreted protein Rv2525c.
J.Struct.Biol., 188, 2014
|
|
2N7O
 
 | | NMR Structure of Peptide PG-990 in DPC micelles | | 分子名称: | Peptide PG-990 | | 著者 | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | 登録日 | 2015-09-16 | | 公開日 | 2015-12-16 | | 最終更新日 | 2017-10-11 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
4KZL
 
 | | Crystal structure of human tankyrase 2 in complex with 4'-fluoro flavone | | 分子名称: | 2-(4-fluorophenyl)-4H-chromen-4-one, GLYCEROL, SULFATE ION, ... | | 著者 | Narwal, M, Haikarainen, T, Lehtio, L. | | 登録日 | 2013-05-30 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of tankyrase inhibiting flavones with increased potency and isoenzyme selectivity.
J.Med.Chem., 56, 2013
|
|
3PC3
 
 | |
4GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 Y108F MUTANT | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | 著者 | Oakley, A, Rossjohn, J, Parker, M. | | 登録日 | 1997-01-20 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Multifunctional role of Tyr 108 in the catalytic mechanism of human glutathione transferase P1-1. Crystallographic and kinetic studies on the Y108F mutant enzyme.
Biochemistry, 36, 1997
|
|
4L0I
 
 | | Tankyrase 2 catalytic domain in complex with ethyl 4-(4-oxo-4H-chromen-2-yl)benzoate | | 分子名称: | SULFATE ION, Tankyrase-2, ZINC ION, ... | | 著者 | Narwal, M, Haikarainen, T, Lehtio, L. | | 登録日 | 2013-05-31 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery of tankyrase inhibiting flavones with increased potency and isoenzyme selectivity.
J.Med.Chem., 56, 2013
|
|
3DCW
 
 | | Use of Carbonic Anhydrase II, IX Active-Site Mimic, for the Purpose of Screening Inhibitors for Possible Anti-Cancer Properties | | 分子名称: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | 著者 | Genis, C, Sippel, K.H, Case, N, Govindasamy, L, Agbandje-Mckenna, M, Mckenna, R. | | 登録日 | 2008-06-04 | | 公開日 | 2009-03-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Design of a carbonic anhydrase IX active-site mimic to screen inhibitors for possible anticancer properties
Biochemistry, 48, 2009
|
|
1L2E
 
 | | Human Kallikrein 6 (hK6) Active Form with benzamidine inhibitor | | 分子名称: | BENZAMIDINE, Kallikrein 6, MAGNESIUM ION | | 著者 | Bernett, M.J, Blaber, S.I, Scarisbrick, I.A, Blaber, M. | | 登録日 | 2002-02-20 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure and biochemical characterization of human kallikrein 6 reveals that a trypsin-like kallikrein is expressed in the central nervous system.
J.Biol.Chem., 277, 2002
|
|
4L0T
 
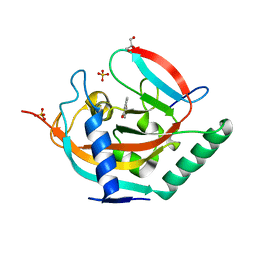 | | Tankyrase 2 in complex with 4'-nitro flavone | | 分子名称: | 2-(4-nitrophenyl)-4H-chromen-4-one, GLYCEROL, SULFATE ION, ... | | 著者 | Narwal, M, Haikarainen, T, Lehtio, L. | | 登録日 | 2013-06-01 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of tankyrase inhibiting flavones with increased potency and isoenzyme selectivity.
J.Med.Chem., 56, 2013
|
|
6HRB
 
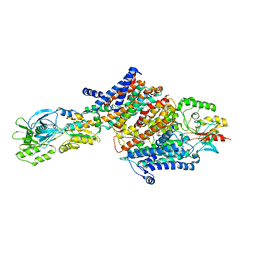 | | Cryo-EM structure of the KdpFABC complex in an E2 inward-facing state (state 2) | | 分子名称: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | 著者 | Stock, C, Hielkema, L, Tascon, I, Wunnicke, D, Oostergetel, G.T, Azkargorta, M, Paulino, C, Haenelt, I. | | 登録日 | 2018-09-26 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structures of KdpFABC suggest a K+transport mechanism via two inter-subunit half-channels.
Nat Commun, 9, 2018
|
|
3PEI
 
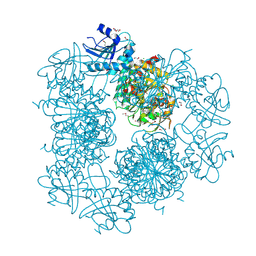 | | Crystal Structure of Cytosol Aminopeptidase from Francisella tularensis | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytosol aminopeptidase, ... | | 著者 | Maltseva, N, Kim, Y, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-10-26 | | 公開日 | 2010-12-01 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure of Cytosol Aminopeptidase from
Francisella tularensis
To be Published
|
|
