4IX5
 
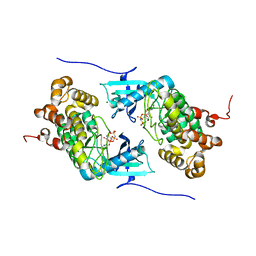 | | Crystal structure of a Stt7 homolog from Micromonas algae in complex with AMP-PNP | | 分子名称: | MAGNESIUM ION, MsStt7d protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Guo, J, Wei, X, Li, M, Pan, X, Chang, W, Liu, Z. | | 登録日 | 2013-01-24 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of the catalytic domain of a state transition kinase homolog from Micromonas algae
Protein Cell, 4, 2013
|
|
6JPF
 
 | |
4EZ5
 
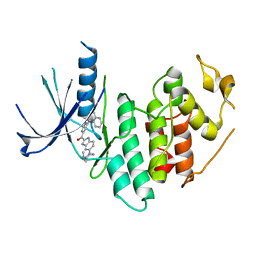 | | CDK6 (monomeric) in complex with inhibitor | | 分子名称: | Cyclin-dependent kinase 6, {5-[4-(dimethylamino)piperidin-1-yl]-1H-imidazo[4,5-b]pyridin-2-yl}[2-(isoquinolin-4-yl)pyridin-4-yl]methanone | | 著者 | Chopra, R, Xu, M. | | 登録日 | 2012-05-02 | | 公開日 | 2013-02-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
1IYB
 
 | |
4IYG
 
 | |
3FJK
 
 | |
6IXD
 
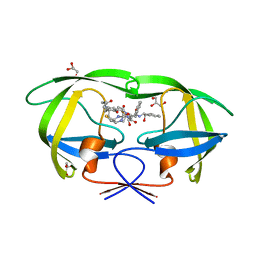 | | X-ray crystal structure of bPI-11 hiv-1 protease complex | | 分子名称: | (4R)-3-[(2S,3S)-3-[2-[4-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]-2,6-dimethyl-phenoxy]ethanoylamino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Adachi, M, Hidaka, K. | | 登録日 | 2018-12-10 | | 公開日 | 2019-05-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Acquired Removability of Aspartic Protease Inhibitors by Direct Biotinylation.
Bioconjug.Chem., 30, 2019
|
|
4ITQ
 
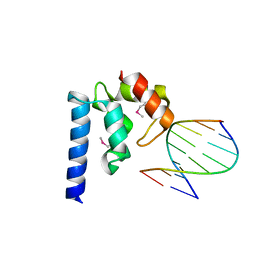 | | Crystal structure of hypothetical protein SCO1480 bound to DNA | | 分子名称: | 5'-D(P*CP*CP*GP*CP*GP*CP*GP*C)-3', 5'-D(P*GP*CP*GP*CP*GP*CP*GP*G)-3', Putative uncharacterized protein SCO1480 | | 著者 | Guarne, A, Nanji, T, Gloyd, M, Swiercz, J.P, Elliot, M.A. | | 登録日 | 2013-01-18 | | 公開日 | 2013-03-27 | | 最終更新日 | 2013-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A novel nucleoid-associated protein specific to the actinobacteria.
Nucleic Acids Res., 41, 2013
|
|
3PPM
 
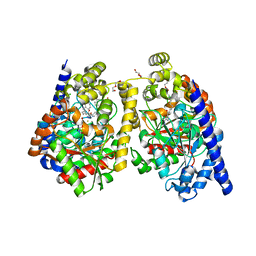 | | Crystal Structure of a Noncovalently Bound alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) to a Humanized Variant of Fatty Acid Amide Hydrolase | | 分子名称: | 1-DODECANOL, 7-phenyl-1-[5-(pyridin-2-yl)-1,3,4-oxadiazol-2-yl]heptan-1-one, CHLORIDE ION, ... | | 著者 | Mileni, M, Han, G.W, Boger, D.L, Stevens, R.C. | | 登録日 | 2010-11-24 | | 公開日 | 2011-11-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Fluoride-mediated capture of a noncovalent bound state of a reversible covalent enzyme inhibitor: X-ray crystallographic analysis of an exceptionally potent alpha-ketoheterocycle inhibitor of fatty acid amide hydrolase.
J.Am.Chem.Soc., 133, 2011
|
|
3FAV
 
 | | Structure of the CFP10-ESAT6 complex from Mycobacterium tuberculosis | | 分子名称: | 6 kDa early secretory antigenic target, ESAT-6-like protein esxB, IMIDAZOLE, ... | | 著者 | Poulsen, C, Holton, S.J, Wilmanns, M, Song, Y.H. | | 登録日 | 2008-11-18 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | WXG100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern.
Plos One, 9, 2014
|
|
6SLB
 
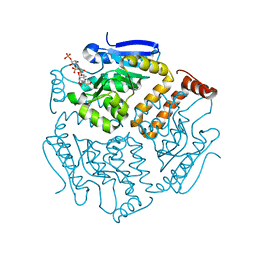 | | Crystal structure of isomerase PaaG with trans-3,4-didehydroadipyl-CoA | | 分子名称: | (~{E})-6-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylsulfanyl]-6-oxidanylidene-hex-3-enoic acid, Enoyl-CoA hydratase/carnithine racemase | | 著者 | Saleem-Batcha, R, Spieker, M, Teufel, R. | | 登録日 | 2019-08-19 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Structural and Mechanistic Basis of an Oxepin-CoA Forming Isomerase in Bacterial Primary and Secondary Metabolism.
Acs Chem.Biol., 14, 2019
|
|
3PIM
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in unusual oxidized form | | 分子名称: | Peptide methionine sulfoxide reductase | | 著者 | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | 登録日 | 2010-11-07 | | 公開日 | 2011-02-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3FBT
 
 | | Crystal structure of a chorismate mutase/shikimate 5-dehydrogenase fusion protein from Clostridium acetobutylicum | | 分子名称: | SULFATE ION, chorismate mutase and shikimate 5-dehydrogenase fusion protein | | 著者 | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-11-19 | | 公開日 | 2008-12-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of a chorismate mutase/shikimate 5-dehydrogenase fusion protein from Clostridium acetobutylicum
To be Published
|
|
6JG1
 
 | | Crystal structure of barley exohydrolaseI wildtype in complex with 4I,4III,4V-S-trithiocellohexaose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Barley exohydrolase I, GLYCEROL, ... | | 著者 | Luang, S, Streltsov, V.A, Hrmova, M. | | 登録日 | 2019-02-13 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
3FKU
 
 | | Crystal structure of influenza hemagglutinin (H5) in complex with a broadly neutralizing antibody F10 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Neutralizing antibody F10, ... | | 著者 | Hwang, W.C, Santelli, E, Stec, B, Wei, G, Cadwell, G, Bankston, L.A, Sui, J, Perez, S, Aird, D, Chen, L.M, Ali, M, Murakami, A, Yammanuru, A, Han, T, Cox, N, Donis, R.O, Liddington, R.C, Marasco, W.A. | | 登録日 | 2008-12-17 | | 公開日 | 2009-02-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4OP4
 
 | | Crystal structure of the catalytic domain of DapE protein from V.cholerea in the Zn bound form | | 分子名称: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, GLYCEROL, ... | | 著者 | Nocek, B, Makowska-Grzyska, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-02-04 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | The Dimerization Domain in DapE Enzymes Is required for Catalysis.
Plos One, 9, 2014
|
|
4J0C
 
 | | tannin acyl hydrolase from Lactobacillus plantarum (native structure) | | 分子名称: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Tannase | | 著者 | Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | 登録日 | 2013-01-30 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
2MHU
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF HUMAN [113CD7] METALLOTHIONEIN-2 IN SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | 分子名称: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | 著者 | Braun, W, Messerle, B.A, Schaeffer, A, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | 登録日 | 1990-05-14 | | 公開日 | 1991-04-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of human [113Cd7]metallothionein-2 in solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
6JGG
 
 | | Crystal structure of barley exohydrolaseI W434F mutant in complex with methyl 2-thio-beta-sophoroside. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | 著者 | Luang, S, Streltsov, V.A, Hrmova, M. | | 登録日 | 2019-02-14 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
4J16
 
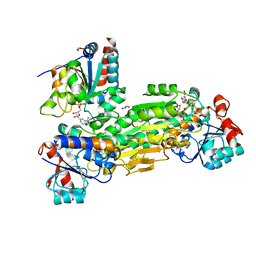 | | Crystal structure of Thermus thermophilus transhydrogenase heterotrimeric complex of the Alpha1 subunit dimer with the NADP binding domain (domain III) of the Beta subunit | | 分子名称: | CHLORIDE ION, GLYCEROL, NAD(P) transhydrogenase subunit beta, ... | | 著者 | Yamaguchi, M, Leung, J, Schurig Briccio, L.A, Gennis, R.B, Stout, C.D. | | 登録日 | 2013-02-01 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Crystal structure analysis of Thermus thermophilus transhydrogenase soluble domains
To be Published
|
|
3PSW
 
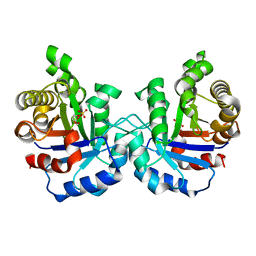 | | Structure of E97Q mutant of TIM from Plasmodium falciparum | | 分子名称: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | 著者 | Samanta, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | 登録日 | 2010-12-02 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Revisiting the mechanism of the triosephosphate isomerase reaction: the role of the fully conserved glutamic acid 97 residue
Chembiochem, 12, 2011
|
|
6F8N
 
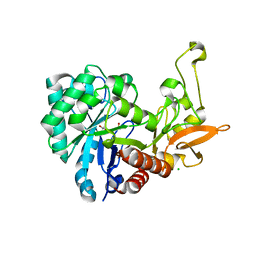 | | Key residues affecting transglycosylation activity in family 18 chitinases - Insights into donor and acceptor subsites | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Madhuprakash, J, Dalhus, B, Swaroopa Rani, T, Podile, A.R, Eijsink, V.G.H, Sorlie, M. | | 登録日 | 2017-12-13 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Key Residues Affecting Transglycosylation Activity in Family 18 Chitinases: Insights into Donor and Acceptor Subsites.
Biochemistry, 57, 2018
|
|
1IT6
 
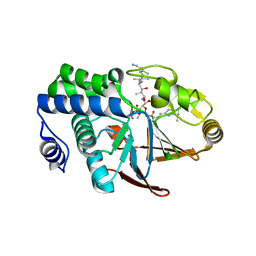 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CALYCULIN A AND THE CATALYTIC SUBUNIT OF PROTEIN PHOSPHATASE 1 | | 分子名称: | CALYCULIN A, MANGANESE (II) ION, SERINE/THREONINE PROTEIN PHOSPHATASE 1 GAMMA (PP1-GAMMA) CATALYTIC SUBUNIT | | 著者 | Kita, A, Matsunaga, S, Takai, A, Kataiwa, H, Wakimoto, T, Fusetani, N, Isobe, M, Miki, K. | | 登録日 | 2002-01-09 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the complex between calyculin A and the catalytic subunit of protein phosphatase 1.
Structure, 10, 2002
|
|
1J2U
 
 | | Creatininase Zn | | 分子名称: | SULFATE ION, ZINC ION, creatinine amidohydrolase | | 著者 | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | 登録日 | 2003-01-11 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
3PTL
 
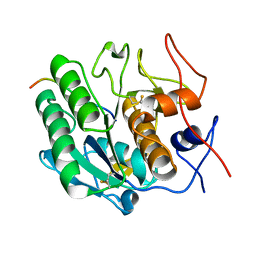 | | Crystal structure of proteinase K inhibited by a lactoferrin nonapeptide, Lys-Gly-Glu-Ala-Asp-Ala-Leu-Ser-Leu-Asp at 1.3 A resolution. | | 分子名称: | 10-mer peptide from Lactoferrin, Proteinase K | | 著者 | Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2010-12-03 | | 公開日 | 2010-12-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal structure of proteinase K inhibited by a lactoferrin nonapeptide, Lys-Gly-Glu-Ala-Asp-Ala-Leu-Ser-Leu-Asp at 1.3 A resolution.
TO BE PUBLISHED
|
|
