6PRV
 
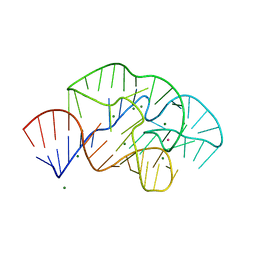 | |
5AC4
 
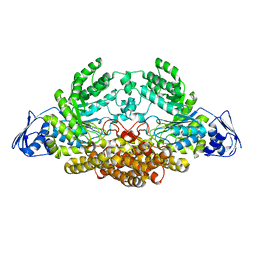 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with GalNAc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-galactopyranose, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | 著者 | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | 登録日 | 2015-08-11 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5FJN
 
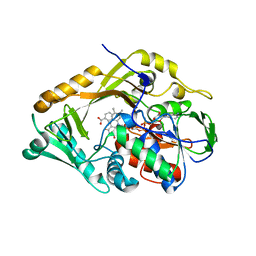 | | Structure of L-Amino acid deaminase from Proteus myxofaciens in complex with anthranilate | | 分子名称: | 2-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID DEAMINASE | | 著者 | Motta, P, Molla, G, Pollegioni, L, Nardini, M. | | 登録日 | 2015-10-11 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-Function Relationships in L-Amino Acid Deaminase, a Flavoprotein Belonging to a Novel Class of Biotechnologically Relevant Enzymes
J.Biol.Chem., 291, 2016
|
|
5JJJ
 
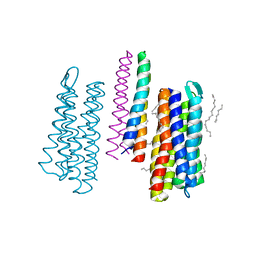 | | Structure of the SRII/HtrII Complex in P64 space group ("U" shape) | | 分子名称: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | 著者 | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Polovinkin, V, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | 登録日 | 2016-04-24 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | New Insights on Signal Propagation by Sensory Rhodopsin II/Transducer Complex.
Sci Rep, 7, 2017
|
|
1AKR
 
 | | G61A OXIDIZED FLAVODOXIN MUTANT | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | 著者 | Mccarthy, A, Walsh, M, Higgins, T. | | 登録日 | 1997-05-27 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
1AKW
 
 | | G61L OXIDIZED FLAVODOXIN MUTANT | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | 著者 | Mccarthy, A, Walsh, M, Higgins, T. | | 登録日 | 1997-05-27 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
5OGQ
 
 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with WRR391 inhibitor | | 分子名称: | ACETATE ION, Cathepsin B-like peptidase (C01 family), ethyl 1-[[(2~{S})-3-(4-hydroxyphenyl)-1-oxidanylidene-1-[[(3~{S})-1-phenyl-5-pyridin-2-ylsulfonyl-pentan-3-yl]amino]propan-2-yl]carbamoyl]piperidine-4-carboxylate | | 著者 | Jilkova, A, Rezacova, P, Brynda, J, Mares, M. | | 登録日 | 2017-07-13 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Druggable Hot Spots in the Schistosomiasis Cathepsin B1 Target Identified by Functional and Binding Mode Analysis of Potent Vinyl Sulfone Inhibitors.
Acs Infect Dis., 2020
|
|
5FJM
 
 | | Structure of L-Amino acid deaminase from Proteus myxofaciens | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID DEAMINASE | | 著者 | Motta, P, Molla, G, Pollegioni, L, Nardini, M. | | 登録日 | 2015-10-11 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Function Relationships in L-Amino Acid Deaminase, a Flavoprotein Belonging to a Novel Class of Biotechnologically Relevant Enzymes
J.Biol.Chem., 291, 2016
|
|
1JD8
 
 | | Solution structure of lactam analogue DapD of HIV gp41 600-612 loop | | 分子名称: | Transmembrane protein gp41 | | 著者 | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | 登録日 | 2001-06-13 | | 公開日 | 2003-07-01 | | 最終更新日 | 2018-10-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
5JMB
 
 | |
6FWJ
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-mannosamine) and alpha-1,2-mannobiose | | 分子名称: | (1~{R},2~{R},3~{R},4~{R},6~{R})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | 著者 | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | 登録日 | 2018-03-06 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
5FUV
 
 | | catalytic domain of Thymidine kinase from Trypanosoma brucei with dThd | | 分子名称: | GLYCEROL, PHOSPHATE ION, THYMDINE KINASE, ... | | 著者 | Timm, J, Valente, M, Castillo-Acosta, V, Balzarini, T, Nettleship, J.E, Rada, H, Wilson, K.S, Gonzalez-Pacanowska, D. | | 登録日 | 2016-01-31 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Cell Cycle Regulation and Novel Structural Features of Thymidine Kinase, an Essential Enzyme in Trypanosoma Brucei.
Mol.Microbiol., 102, 2016
|
|
4L91
 
 | | Crystal structure of Human Hsp90 with X29 | | 分子名称: | 4-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-6-chlorobenzene-1,3-diol, Heat shock protein HSP 90-alpha | | 著者 | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | 登録日 | 2013-06-18 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7QW4
 
 | | Pden_5119 protein | | 分子名称: | NADPH-dependent FMN reductase | | 著者 | Kryl, M, Sedlacek, V. | | 登録日 | 2022-01-24 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Insight into Catalysis by the Flavin-Dependent NADH Oxidase (Pden_5119) of Paracoccus denitrificans .
Int J Mol Sci, 24, 2023
|
|
5JQQ
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis - apo form | | 分子名称: | GLYCEROL, Glucosyl-3-phosphoglycerate synthase | | 著者 | Albesa-Jove, D, Urresti, S, Gest, P.M, van der Woerd, M, Jackson, M, Guerin, M.E. | | 登録日 | 2016-05-05 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis - apo form
To Be Published
|
|
1AAP
 
 | | X-RAY CRYSTAL STRUCTURE OF THE PROTEASE INHIBITOR DOMAIN OF ALZHEIMER'S AMYLOID BETA-PROTEIN PRECURSOR | | 分子名称: | ALZHEIMER'S DISEASE AMYLOID A4 PROTEIN | | 著者 | Hynes, T.R, Randal, M, Kennedy, L.A, Eigenbrot, C, Kossiakoff, A.A. | | 登録日 | 1990-09-14 | | 公開日 | 1991-10-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray crystal structure of the protease inhibitor domain of Alzheimer's amyloid beta-protein precursor.
Biochemistry, 29, 1990
|
|
8D45
 
 | |
5JRB
 
 | | Rad52(1-212) K102A/K133A/E202A mutant | | 分子名称: | DNA repair protein RAD52 homolog | | 著者 | Saotome, M, Saito, K, Kurumizaka, H, Kagawa, W. | | 登録日 | 2016-05-06 | | 公開日 | 2016-08-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.405 Å) | | 主引用文献 | Structure of the human DNA-repair protein RAD52 containing surface mutations.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5JRO
 
 | | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form | | 分子名称: | FMN-dependent NADH-azoreductase, GLYCEROL | | 著者 | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-05-06 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form
To Be Published
|
|
8DF5
 
 | | SARS-CoV-2 Beta RBD in complex with human ACE2 and S304 Fab and S309 Fab | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | McCallum, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2022-06-21 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution.
Science, 377, 2022
|
|
5A69
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with Gal-PUGNAc | | 分子名称: | N-ACETYL-BETA-D-GLUCOSAMINIDASE, [(Z)-[(3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-ylidene]amino] N-phenylcarbamate | | 著者 | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | 登録日 | 2015-06-24 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5UPY
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria Monocytogenes in the complex with IMP and Q21 | | 分子名称: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-04 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria Monocytogenes in the complex with IMP and Q21
To Be Published
|
|
6FYH
 
 | | Disulfide between ubiquitin G76C and the E3 HECT ligase Huwe1 | | 分子名称: | E3 ubiquitin-protein ligase HUWE1, Polyubiquitin-B, SULFATE ION, ... | | 著者 | Jaeckl, M, Hartmann, M.D, Wiesner, S. | | 登録日 | 2018-03-12 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.906 Å) | | 主引用文献 | beta-Sheet Augmentation Is a Conserved Mechanism of Priming HECT E3 Ligases for Ubiquitin Ligation.
J. Mol. Biol., 430, 2018
|
|
1JAR
 
 | | Solution structure of lactam analogue (DDab)of HIV gp41 600-612 loop. | | 分子名称: | DDab: (ACE)IWGDSGKLI(DAB)TTA ANALOGUE OF HIV GP41 | | 著者 | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | 登録日 | 2001-05-31 | | 公開日 | 2003-07-01 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
5JTS
 
 | | Structure of a beta-1,4-mannanase, SsGH134. | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | 登録日 | 2016-05-09 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
