6YU3
 
 | | Crystal structure of MhsT in complex with L-phenylalanine | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, PHENYLALANINE, ... | | 著者 | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | 登録日 | 2020-04-25 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
8OVY
 
 | | Structure of analogue of superfolded GFP | | 分子名称: | Green fluorescent protein | | 著者 | Dunkelmann, D, Fiedler, M, Bellini, D, Alvira, C.P, Chin, J.W. | | 登録日 | 2023-04-26 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.537 Å) | | 主引用文献 | Adding alpha , alpha-disubstituted and beta-linked monomers to the genetic code of an organism.
Nature, 625, 2024
|
|
8OI1
 
 | | Yeast 20S proteasome in complex with a photoswitchable cepafungin derivative (transCep4) | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | 著者 | Morstein, J, Amatuni, A, Schuster, A, Kuttenlochner, W, Ko, T, Groll, M, Adibekian, A, Renata, H, Trauner, D.H. | | 登録日 | 2023-03-21 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Optical Control of Proteasomal Protein Degradation with a Photoswitchable Lipopeptide.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6ZAU
 
 | | Damage-free nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) determined by serial femtosecond rotation crystallography | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | 著者 | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | 登録日 | 2020-06-05 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6YWO
 
 | | CutA in complex with A3 RNA | | 分子名称: | CHLORIDE ION, CutA, MAGNESIUM ION, ... | | 著者 | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | 登録日 | 2020-04-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2020-09-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
4OBQ
 
 | | MAP4K4 in complex with inhibitor (compound 31), N-[3-(4-AMINOQUINAZOLIN-6-YL)-5-FLUOROPHENYL]-2-(PYRROLIDIN-1-YL)ACETAMIDE | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Mitogen-activated protein kinase kinase kinase kinase 4, ... | | 著者 | Harris, S.F, Wu, P, Coons, M. | | 登録日 | 2014-01-07 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Discovery of Selective 4-Amino-pyridopyrimidine Inhibitors of MAP4K4 Using Fragment-Based Lead Identification and Optimization.
J.Med.Chem., 57, 2014
|
|
5KK4
 
 | | Crystal Structure of the Plant Defensin NsD7 bound to Phosphatidic Acid | | 分子名称: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 1,2-ETHANEDIOL, ACETATE ION, ... | | 著者 | Kvansakul, M, Hulett, M.D, Lay, F.T. | | 登録日 | 2016-06-21 | | 公開日 | 2016-10-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Binding of phosphatidic acid by NsD7 mediates the formation of helical defensin-lipid oligomeric assemblies and membrane permeabilization.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8Q3Z
 
 | |
2Z7F
 
 | |
7SUU
 
 | |
8Q3Y
 
 | |
6ZEA
 
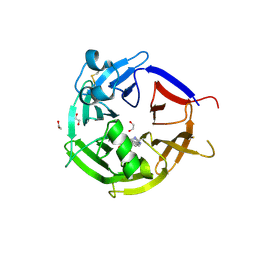 | | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline | | 分子名称: | (1~{R})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, (1~{S})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, 1,2-ETHANEDIOL, ... | | 著者 | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | 登録日 | 2020-06-16 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline
To Be Published
|
|
7SX6
 
 | |
6ZEI
 
 | | Structure of PP1-IRSp53 S455E chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | 分子名称: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | 著者 | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | 登録日 | 2020-06-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
7SX7
 
 | |
6Z1Q
 
 | | MAP3K14 (NIK) in complex with DesF-3R/4076 | | 分子名称: | DesF-3R/4076, Mitogen-activated protein kinase kinase kinase 14 | | 著者 | Jacoby, E, van Vlijmen, H, Querolle, O, Stansfield, I, Meerpoel, L, Versele, M, Hynd, G, Attar, R. | | 登録日 | 2020-05-14 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | FEP+ calculations predict a stereochemical SAR switch for first-in-class indoline NIK inhibitors for multiple myeloma
Future Drug Discov, 2, 2020
|
|
2Z8U
 
 | |
8Q1K
 
 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | 著者 | Roske, Y, Daumke, O, Damme, M. | | 登録日 | 2023-07-31 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
6Z20
 
 | | Structure of the EC2 domain of CD9 in complex with nanobody 4C8 | | 分子名称: | CD9 antigen, CHLORIDE ION, GLYCEROL, ... | | 著者 | Oosterheert, W, Manshande, J, Pearce, N.M, Lutz, M, Gros, P. | | 登録日 | 2020-05-14 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
3UUB
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' mutant reduced in solution | | 分子名称: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | 著者 | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | 登録日 | 2011-11-28 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5KP2
 
 | | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae cocrystallized with octanoyl-CoA: hydrolzed ligand | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, COENZYME A, OCTANOIC ACID (CAPRYLIC ACID) | | 著者 | Hou, J, Zheng, H, Grabowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-07-01 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae soaked with octanoyl-CoA: hydrolzed ligand
To Be Published
|
|
8PVK
 
 | | Chaetomium thermophilum pre-60S State 5 - pre-5S rotation - L1 inward - composite structure | | 分子名称: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVL
 
 | | Chaetomium thermophilum pre-60S State 7 - pre-5S rotation lacking Utp30/ITS2 - composite structure | | 分子名称: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.19 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
5KR3
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-06 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5HS3
 
 | | Human thymidylate synthase complexed with dUMP and 3-amino-2-benzoyl-4-methylthieno[2,3-b]pyridin-6-ol | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 3-amino-2-benzoyl-4-methylthieno[2,3-b]pyridin-6-ol, Thymidylate synthase | | 著者 | Chen, D, Almqvist, H, Axelsson, H, Jafari, R, Mateus, A, Haraldsson, M, Larsson, A, Artursson, P, Molina, D.M, Lundback, T, Nordlund, P. | | 登録日 | 2016-01-25 | | 公開日 | 2016-02-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.103 Å) | | 主引用文献 | CETSA screening identifies known and novel thymidylate synthase inhibitors and slow intracellular activation of 5-fluorouracil
Nat Commun, 7, 2016
|
|
