6M4M
 
 | | X-ray crystal structure of the E249Q mutan of alpha-amylase I and maltohexaose complex from Eisenia fetida | | Descriptor: | Alpha-amylase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M4V
 
 | | Crystal structure of MBP fused split FKBP in complex with rapamycin | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
1UYL
 
 | | Structure-Activity Relationships in purine-based inhibitor binding to HSP90 isoforms | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
6M64
 
 | | Crystal structure of SMAD2 in complex with CBP | | Descriptor: | CBP, Mothers against decapentaplegic homolog 2 | | Authors: | Miyazono, K, Ito, T, Wada, H, Tanokura, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-beta signaling.
Sci.Signal., 13, 2020
|
|
7SKH
 
 | |
7A8T
 
 | | Crystal structure of sarcomeric protein FATZ-1 (mini-FATZ-1 construct) in complex with rod domain of alpha-actinin-2 | | Descriptor: | Alpha-actinin-2, Myozenin-1 | | Authors: | Sponga, A, Arolas, J.L, Rodriguez Chamorro, A, Mlynek, G, Hollerl, E, Schreiner, C, Pedron, M, Kostan, J, Ribeiro, E.A, Djinovic-Carugo, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with alpha-actinin.
Sci Adv, 7, 2021
|
|
1V07
 
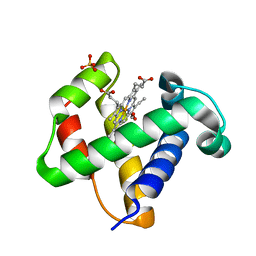 | | CRYSTAL STRUCTURE OF ThrE11Val mutant of THE NERVE TISSUE MINI-HEMOGLOBIN FROM THE NEMERTEAN WORM CEREBRATULUS LACTEUS | | Descriptor: | NEURAL HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Ascenzi, P, Geuens, E, Dewilde, S, Moens, L, Bolognesi, M, Riggs, A, Hale, A, Deng, P, Nienhaus, G.U, Olson, J.S, Nienhaus, K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thr-E11 Regulates O2 Affinity in Cerebratulus Lacteus Mini-Hemoglobin
J.Biol.Chem., 279, 2004
|
|
6UYU
 
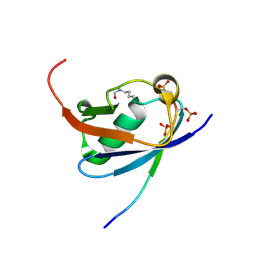 | | Crystal structure of K45-acetylated SUMO1 in complex with phosphorylated PML-SIM | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Wahba, H.M, Gagnon, C, Mascle, X.H, Lussier-Price, M, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-11-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.
Structure, 28, 2020
|
|
1UY6
 
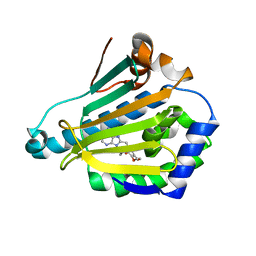 | | Human Hsp90-alpha with 9-Butyl-8-(3,4,5-trimethoxy-benzyl)-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(3,4,5-TRIMETHOXYBENZYL)-9H-PURIN-6-AMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
6UOF
 
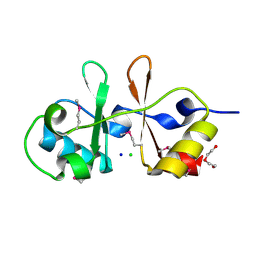 | | 1.2 Angstrom Resolution Crystal Structure of CBS Domains of Transcriptional Regulator from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-10-14 | | Release date: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 1.2 Angstrom Resolution Crystal Structure of CBS Domains of Transcriptional Regulator from Streptococcus pneumoniae
To Be Published
|
|
6IX8
 
 | | The structure of LepI C52A in complex with SAM and its substrate analogue | | Descriptor: | (1R,2R,4aS,8S,8aR)-2,8-dimethyl-5'-phenyl-4a,5,6,7,8,8a-hexahydro-2H,2'H-spiro[naphthalene-1,3'-pyridine]-2',4'(1'H)-dione, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
6LL6
 
 | | Crsyal structure of EcFtsZ (residues 11-316) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yoshizawa, T, Fujita, J, Terakada, H, Ozawa, M, Kuroda, N, Tanaka, S, Uehara, R, Matsumura, H. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the cell-division protein FtsZ from Klebsiella pneumoniae and Escherichia coli.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7DVE
 
 | | Crystal structure of FAD-dependent C-glycoside oxidase | | Descriptor: | 6'''-hydroxyparomomycin C oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Senda, M, Watanabe, S, Kumano, T, Kobayashi, M, Senda, T. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | FAD-dependent C -glycoside-metabolizing enzymes in microorganisms: Screening, characterization, and crystal structure analysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6URB
 
 | |
6M3L
 
 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(nonspecific) complex | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*CP*GP*AP*TP*TP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*GP*CP*G)-3'), RE_R_Pab1 domain-containing protein | | Authors: | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
6V36
 
 | |
6M4L
 
 | | X-ray crystal structure of the E249Q mutant of alpha-amylase I from Eisenia fetida | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1I0F
 
 | | 1.6 A STRUCTURE OF THE A-DECAMER GCGTATACGC WITH A SINGLE 2'-O-AMINOOXYETHYL THYMINE IN PLACE OF T6, BA-FORM | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(127)P*AP*CP*GP*C)-3', BARIUM ION | | Authors: | Tereshko, V, Wilds, C.J, Minasov, G, Prakash, T.P, Maier, M.A, Howard, A, Wawrzak, Z, Manoharan, M, Egli, M. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments.
Nucleic Acids Res., 29, 2001
|
|
6M4U
 
 | | Crystal structure of FKBP-FRB T2098L mutant in complex with rapamycin | | Descriptor: | CHLORIDE ION, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
6ZOM
 
 | | Oxidized thioredoxin 1 from the anaerobic bacteria Desulfovibrio vulgaris Hildenborough | | Descriptor: | Thioredoxin | | Authors: | Garcin, E, Bornet, O, Nouailler, M, Pieulle, L, Guerlesquin, F, Sebban-Kreuzer, C. | | Deposit date: | 2020-07-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Glutamate optimizes enzymatic activity under high hydrostatic pressure in Desulfovibrio species: effects on the ubiquitous thioredoxin system.
Extremophiles, 25, 2021
|
|
6UTQ
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with cadmium | | Descriptor: | ATP-dependent sacrificial sulfur transferase LarE, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Fellner, M, Huizenga, K, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystallographic characterization of a tri-Asp metal-binding site at the three-fold symmetry axis of LarE.
Sci Rep, 10, 2020
|
|
1I0G
 
 | | 1.45 A STRUCTURE OF THE A-DECAMER GCGTATACGC WITH A SINGLE 2'-O-FLUOROETHYL THYMINE IN PLACE OF T6, MEDIUM NA-SALT | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(125)P*AP*CP*GP*C)-3', SODIUM ION | | Authors: | Tereshko, V, Wilds, C.J, Minasov, G, Prakash, T.P, Maier, M.A, Howard, A, Wawrzak, Z, Manoharan, M, Egli, M. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments.
Nucleic Acids Res., 29, 2001
|
|
1I0J
 
 | | 1.06 A STRUCTURE OF THE A-DECAMER GCGTATACGC WITH A SINGLE 2'-O-METHYL-3'-METHYLENEPHOSPHONATE (T23) THYMINE IN PLACE OF T6, HIGH CS-SALT | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(T23)P*AP*CP*GP*C)-3', CESIUM ION | | Authors: | Tereshko, V, Wilds, C.J, Minasov, G, Prakash, T.P, Maier, M.A, Howard, A, Wawrzak, Z, Manoharan, M, Egli, M. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments.
Nucleic Acids Res., 29, 2001
|
|
1UUE
 
 | | a-SPECTRIN SH3 DOMAIN (V44T, D48G MUTANT) | | Descriptor: | SPECTRIN ALPHA CHAIN | | Authors: | Vega, M.C, Fernandez, A, Wilmanns, M, Serrano, L. | | Deposit date: | 2003-12-18 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Solvation in Protein Folding Analysis: Combination of Theoretical and Experimental Approaches
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1I0K
 
 | | 1.05 A STRUCTURE OF THE A-DECAMER GCGTATACGC WITH A SINGLE 2'-O-METHYL-[TRI(OXYETHYL)] THYMINE IN PLACE OF T6, MEDIUM CS-SALT | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(126)P*AP*CP*GP*C)-3', CESIUM ION | | Authors: | Tereshko, V, Wilds, C.J, Minasov, G, Prakash, T.P, Maier, M.A, Howard, A, Wawrzak, Z, Manoharan, M, Egli, M. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments.
Nucleic Acids Res., 29, 2001
|
|
