5ESZ
 
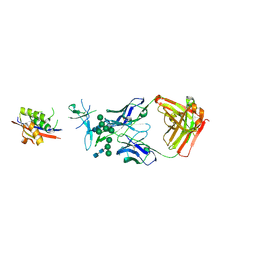 | | Crystal Structure of Broadly Neutralizing Antibody CH04, Isolated from Donor CH0219, in Complex with Scaffolded Trimeric HIV-1 Env V1V2 Domain from the Clade AE Strain A244 | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase,Envelope glycoprotein gp160, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH04 Heavy Chain, ... | | Authors: | Gorman, J, Yang, M, Kwong, P.D. | | Deposit date: | 2015-11-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.191 Å) | | Cite: | Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5ESV
 
 | | Crystal Structure of Broadly Neutralizing Antibody CH03, Isolated from Donor CH0219, in Complex with Scaffolded Trimeric HIV-1 Env V1V2 Domain from the Clade C Superinfecting Strain of Donor CAP256. | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase,Envelope glycoprotein gp160, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorman, J, Yang, M, Kwong, P.D. | | Deposit date: | 2015-11-17 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8OYT
 
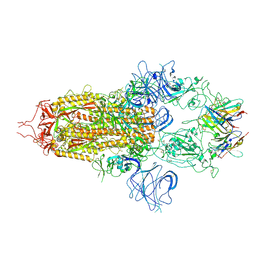 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | Authors: | Weckener, M, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
5SAK
 
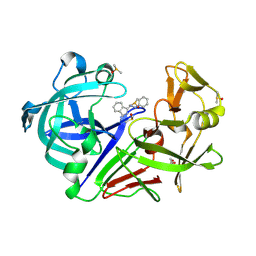 | | Endothiapepsin in complex with compound FU5-1 | | Descriptor: | (1Z,3Z)-3-(2-phenylhydrazinylidene)-2,3-dihydro-1H-isoindol-1-imine, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAQ
 
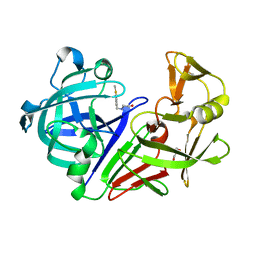 | | Endothiapepsin in complex with compound FU58-3 | | Descriptor: | 2-({[4-(methylsulfanyl)phenyl]methyl}amino)ethan-1-ol, Endothiapepsin, GLYCEROL | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAR
 
 | | Endothiapepsin in complex with compound FU290-1 | | Descriptor: | (1,4-phenylene)bis(methylene) dicarbamimidothioate, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7JVM
 
 | |
5SAO
 
 | | Endothiapepsin in complex with compound FU58-1 | | Descriptor: | 1,2-ETHANEDIOL, 6-[(8R)-2-({[(3,5-dimethyl-1,2-oxazol-4-yl)methyl](methyl)amino}methyl)-6,7-dihydropyrazolo[1,5-a]pyrazin-5(4H)-yl]pyrimidin-4-amine, Endothiapepsin, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAL
 
 | | Endothiapepsin in complex with compound FU5-2 | | Descriptor: | (1Z)-1-imino-1H-isoindol-3-amine, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAT
 
 | | Endothiapepsin in complex with compound FU66-1 | | Descriptor: | Endothiapepsin, GLYCEROL, N-cyclopropyl-6-(furan-2-yl)-2-hydroxy-N-[(pyridin-2-yl)methyl]pyridine-3-carboxamide | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2G51
 
 | | anomalous substructure of trypsin (p1) | | Descriptor: | CHLORIDE ION, Trypsin | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5SAN
 
 | | Endothiapepsin in complex with compound FU5-4 | | Descriptor: | (2Z)-2-(3-amino-1H-isoindol-1-ylidene)hydrazine-1-carboximidamide, Endothiapepsin, GLYCEROL | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAM
 
 | | Endothiapepsin in complex with compound FU5-3 | | Descriptor: | 3-amino-1H-isoindol-1-one, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAS
 
 | | Endothiapepsin in complex with compound FU290-2 | | Descriptor: | (1R)-1-(4-chlorophenyl)ethyl carbamimidothioate, BROMIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAP
 
 | | Endothiapepsin in complex with compound FU58-2 | | Descriptor: | (8S)-5-(2-aminopyrimidin-4-yl)-N-[2-(dimethylamino)ethyl]-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazine-2-carboxamide, 1,2-ETHANEDIOL, Endothiapepsin, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5LX7
 
 | | Cys-Gly dipeptidase GliJ mutant D38N | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dipeptidase, FE (III) ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
7NO8
 
 | | Structure of the mature RSV CA lattice: Group I, pentamer-hexamer interface, class 1"6 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
6NYC
 
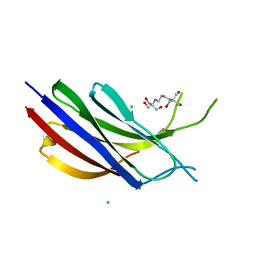 | | Munc13-1 C2B-domain, calcium free | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Munc13-1 | | Authors: | Tomchick, D.R, Rizo, J, Machius, M, Lu, J. | | Deposit date: | 2019-02-11 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis.
Nat. Struct. Mol. Biol., 17, 2010
|
|
7NOM
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 4'Beta | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOG
 
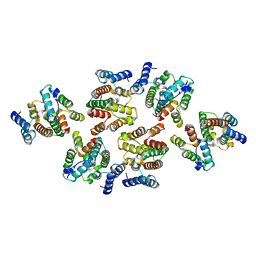 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 4'5 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO0
 
 | | Structure of the mature RSV CA lattice: T=1 CA icosahedron | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOB
 
 | | Structure of the mature RSV CA lattice: Group II, hexamer-hexamer interface, class 2'6 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOI
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 3'Alpha | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
5EZR
 
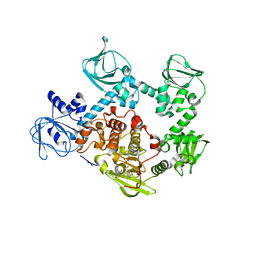 | | Crystal Structure of PVX_084705 bound to compound | | Descriptor: | CHLORIDE ION, N-[5-(3-{2-[(cyclopropylmethyl)amino]pyrimidin-4-yl}-7-[(dimethylamino)methyl]-6-methylimidazo[1,2-a]pyridin-2-yl)-2-fluorophenyl]methanesulfonamide, cGMP-dependent protein kinase, ... | | Authors: | El Bakkouri, M, Amani, M, Walker, J.R, Osborne, S, Large, J.M, Birchall, K, Bouloc, N, Smiljanic-Hurley, E, Wheldon, M, Harding, D.J, Merritt, A.T, Ansell, K.H, Coombs, P.J, Kettleborough, C.A, Stewart, B.L, Bowyer, P.W, Gutteridge, W.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Baker, D.A, Hui, R, Loppnau, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-26 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PVX_084705 bound to compound
To Be Published
|
|
7NO3
 
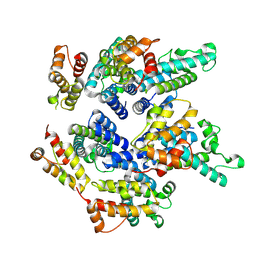 | | Structure of the mature RSV CA lattice: pentamer derived from polyhedral VLPs | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
