7EAP
 
 | | Crystal structure of IpeA-XXXG complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Matsuzawa, T, Watanabe, M, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis for the catalytic mechanism of the glycoside hydrolase family 3 isoprimeverose-producing oligoxyloglucan hydrolase from Aspergillus oryzae.
Febs Lett., 596, 2022
|
|
3EM6
 
 | | Crystal structure of I50L/A71V mutant of hiv-1 protease in complex with inhibitor darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Royer, C.J, King, N.M, Prabu-Jeyabalan, M, Ng, C, Nalivaika, E.A, Schiffer, C.A. | | Deposit date: | 2008-09-23 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural studies on atazanavir specific I50L drug-resistant HIV-1 protease mutant.
To be Published
|
|
7EAW
 
 | | Trehalase of Arabidopsis thaliana acid mutant -D380A trehalose complex | | Descriptor: | GLYCEROL, Trehalase, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
7Y00
 
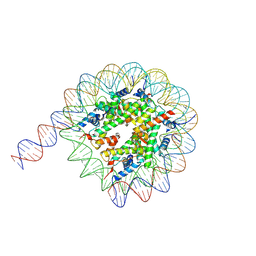 | | Cryo-EM structure of the nucleosome containing 169 base-pair DNA with a p53 target sequence | | Descriptor: | DNA (169-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7XZC
 
 | |
3E6Y
 
 | | Structure of 14-3-3 in complex with the differentiation-inducing agent Cotylenin A | | Descriptor: | 14-3-3-like protein C, CHLORIDE ION, Cotylenin A, ... | | Authors: | Ottmann, C, Weyand, M, Wittinghofer, A, Oecking, C. | | Deposit date: | 2008-08-17 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural rationale for selective stabilization of anti-tumor interactions of 14-3-3 proteins by cotylenin A
J.Mol.Biol., 386, 2009
|
|
3E7N
 
 | | Crystal structure of d-ribose high-affinity transport system from salmonella typhimurium lt2 | | Descriptor: | 1,2-ETHANEDIOL, D-ribose high-affinity transport system | | Authors: | Nocek, B, Maltseva, N, Gu, M, Joachimiak, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-18 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of d-ribose high-affinity transport system from salmonella typhimurium lt2
To be Published
|
|
3E4B
 
 | | Crystal structure of AlgK from Pseudomonas fluorescens WCS374r | | Descriptor: | AlgK, CHLORIDE ION, GLYCEROL | | Authors: | Keiski, C.-L, Harwich, M, Jain, S, Neculai, A.M, Yip, P, Robinson, H, Whitney, J.C, Burrows, L.L, Ohman, D.E, Howell, P.L. | | Deposit date: | 2008-08-11 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin.
Structure, 18, 2010
|
|
7EFI
 
 | |
7EFH
 
 | | RNA kink-turn motif | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*AP*AP*GP*AP*AP*CP*CP*GP*GP*GP*GP*AP*GP*CP*C)-3') | | Authors: | Kondo, J, Nagashima, M. | | Deposit date: | 2021-03-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RNA kink-turn motif
To Be Published
|
|
3E7L
 
 | | Crystal structure of sigma54 activator NtrC4's DNA binding domain | | Descriptor: | Transcriptional regulator (NtrC family), ZINC ION | | Authors: | Batchelor, J.D, Doucleff, M, Lee, C.-J, Matsubara, K, De Carlo, S, Heideker, J, Lamers, M.M, Pelton, J.G, Wemmer, D.E. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation.
J.Mol.Biol., 384, 2008
|
|
7Y8Q
 
 | | Amyloid-beta assemblage on GM1-containing membranes | | Descriptor: | Amyloid-beta protein 40 | | Authors: | Yagi-Utsumi, M, Itoh, S.G, Okumura, H, Yanagisawa, K, Kato, K, Nishimura, K. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The Double-Layered Structure of Amyloid-beta Assemblage on GM1-Containing Membranes Catalytically Promotes Fibrillization.
Acs Chem Neurosci, 14, 2023
|
|
7EI2
 
 | | Structure of human NNMT in complex with macrocyclic peptide 8 | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide 8 | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
7EIF
 
 | | Crystal structure of GAS41 YEATS domain | | Descriptor: | GLYCEROL, YEATS domain-containing protein 4 | | Authors: | Kikuchi, M, Umehara, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | GAS41 promotes H2A.Z deposition through recognition of the N terminus of histone H3 by the YEATS domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3EEQ
 
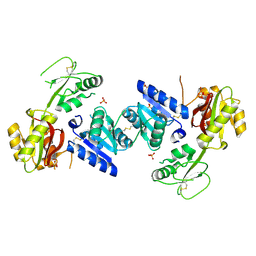 | | Crystal structure of a putative cobalamin biosynthesis protein G homolog from Sulfolobus solfataricus | | Descriptor: | SULFATE ION, putative Cobalamin biosynthesis protein G homolog | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-05 | | Release date: | 2008-09-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative cobalamin biosynthesis protein G homolog from Sulfolobus solfataricus
To be Published
|
|
7ELK
 
 | |
7EFG
 
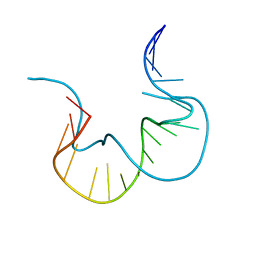 | | RNA kink-turn motif | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*AP*AP*GP*AP*AP*CP*CP*GP*GP*GP*GP*AP*GP*CP*C)-3') | | Authors: | Kondo, J, Nagashima, M. | | Deposit date: | 2021-03-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RNA kink-turn motif
To Be Published
|
|
3ELL
 
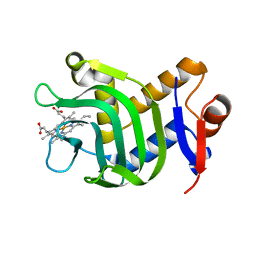 | | Structure of the hemophore from Pseudomonas aeruginosa (HasAp) | | Descriptor: | HasAp (Heme acquisition protein HasAp), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schonbrunn, E, Rivera, M. | | Deposit date: | 2008-09-22 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of the hemophore HasAp from Pseudomonas aeruginosa: NMR spectroscopy reveals protein-protein interactions between Holo-HasAp and hemoglobin.
Biochemistry, 48, 2009
|
|
3EIF
 
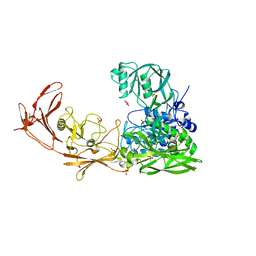 | | 1.9 angstrom crystal structure of the active form of the C5a peptidase from Streptococcus pyogenes (ScpA) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Cooney, J.C, Kagawa, T.F, O'Connell, M.R, Paoli, M, Mouat, P, O'Toole, P.W. | | Deposit date: | 2008-09-15 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Model for Substrate Interactions in C5a Peptidase from Streptococcus pyogenes: A 1.9 A Crystal Structure of the Active Form of ScpA
J.Mol.Biol., 386, 2009
|
|
7EKU
 
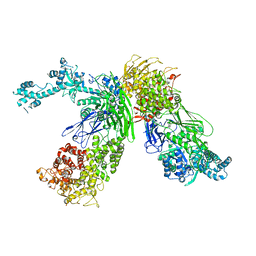 | |
6K8M
 
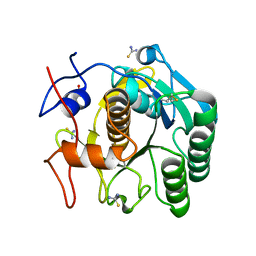 | |
3B1X
 
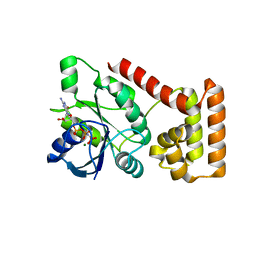 | | Crystal structure of an S. thermophilus NFeoB E66A mutant bound to GMPPNP | | Descriptor: | Ferrous iron uptake transporter protein B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-07-15 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | A suite of Switch I and Switch II mutant structures from the G-protein domain of FeoB
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3B0P
 
 | | tRNA-dihydrouridine synthase from Thermus thermophilus | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-12 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7EG5
 
 | |
3B4M
 
 | | Crystal Structure of Human PABPN1 RRM | | Descriptor: | Polyadenylate-binding protein 2 | | Authors: | Ge, H, Zhou, D, Teng, M, Niu, L. | | Deposit date: | 2007-10-24 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure and possible dimerization of the single RRM of human PABPN1
Proteins, 71, 2008
|
|
