7FNZ
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07F12 from the F2X-Universal Library | | Descriptor: | 2-[(2-amino-2-oxoethyl)sulfanyl]-N-(4-fluorophenyl)acetamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FO7
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07H05 from the F2X-Universal Library | | Descriptor: | (3R)-N-cyclohexylpiperidine-3-carboxamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FO0
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07G02 from the F2X-Universal Library | | Descriptor: | 2-(4-fluorophenoxy)-N-(2-methoxyethyl)acetamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FOG
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08B02 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N~2~-[(1R)-1-(2,4-difluorophenyl)ethyl]-N-ethylglycinamide, N~2~-[(1S)-1-(2,4-difluorophenyl)ethyl]-N-ethylglycinamide, ... | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
5Y4B
 
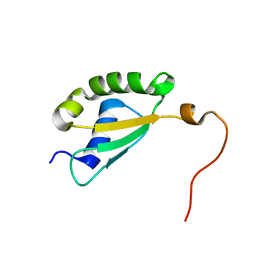 | | Solution structure of yeast Fra2 | | Descriptor: | BolA-like protein 2 | | Authors: | Tang, Y.J, Chi, C.B, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-03 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J. Mol. Biol., 430, 2018
|
|
7FOA
 
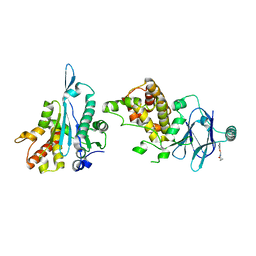 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07H07 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N-cyclopropyl-2-(3-fluorophenoxy)acetamide, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
6QTA
 
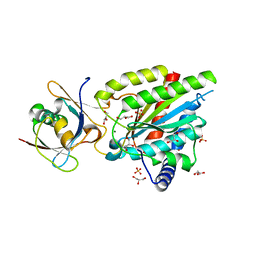 | | Crystal structure of Rea1-MIDAS/Rsa4-UBL complex from Chaetomium thermophilum | | Descriptor: | GLYCEROL, MAGNESIUM ION, Midasin,Midasin, ... | | Authors: | Ahmed, Y.L, Thoms, M, Hurt, E, Sinning, I. | | Deposit date: | 2019-02-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Rea1-MIDAS bound to its ribosome assembly factor ligands resembling integrin-ligand-type complexes.
Nat Commun, 10, 2019
|
|
8RWK
 
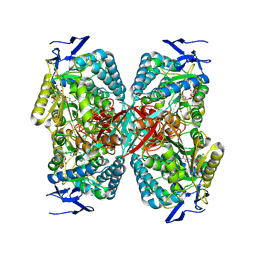 | | cryoEM structure of the central Ald4 filament determined by FilamentID | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium-activated aldehyde dehydrogenase, mitochondrial | | Authors: | Hugener, J, Xu, J, Wettstein, R, Ioannidi, L, Velikov, D, Wollweber, F, Henggeler, A, Matos, J, Pilhofer, M. | | Deposit date: | 2024-02-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | FilamentID reveals the composition and function of metabolic enzyme polymers during gametogenesis.
Cell, 187, 2024
|
|
7FOM
 
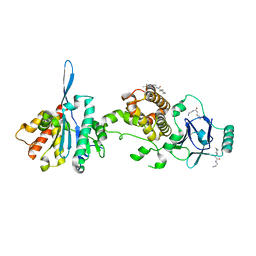 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08C04 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, DIMETHYL SULFOXIDE, N-(2-aminoethyl)-2-cyclohexylacetamide, ... | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FOH
 
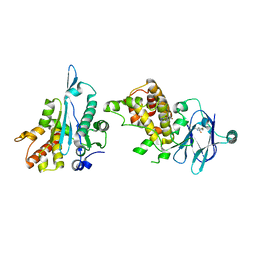 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08B05 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N-[2-(dimethylamino)ethyl]-4-fluorobenzamide, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
8RWJ
 
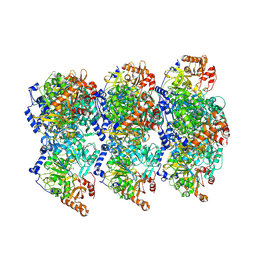 | | cryoEM structure of Acs1 filament determined by FilamentID | | Descriptor: | Acetyl-coenzyme A synthetase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate | | Authors: | Hugener, J, Xu, J, Wettstein, R, Ioannidi, L, Velikov, D, Wollweber, F, Henggeler, A, Matos, J, Pilhofer, M. | | Deposit date: | 2024-02-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | FilamentID reveals the composition and function of metabolic enzyme polymers during gametogenesis.
Cell, 187, 2024
|
|
6QMQ
 
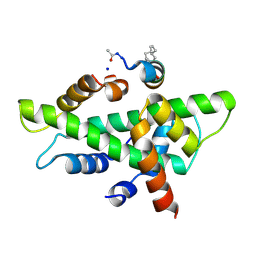 | | NF-YB/C Heterodimer in Complex with NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | Nuclear transcription factor Y subunit alpha, Nuclear transcription factor Y subunit beta, Nuclear transcription factor Y subunit gamma, ... | | Authors: | Kiehstaller, S, Jeganathan, S, Pearce, N.M, Wendt, M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2019-02-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Constrained Peptides with Fine-Tuned Flexibility Inhibit NF-Y Transcription Factor Assembly.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
7FOW
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08E04 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N-(2-hydroxyethyl)-N'-(3-methylphenyl)thiourea, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
6QTS
 
 | |
7FP4
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08G02 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, methyl N-methyl-N-(thiophene-3-carbonyl)glycinate | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FOO
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08C06 from the F2X-Universal Library | | Descriptor: | 2-azanyl-1-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl)ethanone, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FPA
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P09C01 from the F2X-Universal Library | | Descriptor: | 2,4-dichloro-N-ethyl-N-(2-hydroxyethyl)benzamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FNH
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07C05 from the F2X-Universal Library | | Descriptor: | 4-amino-N-[(2R)-1-hydroxybutan-2-yl]benzamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FPG
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P09F06 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, [4-(difluoromethoxy)phenyl]methanol | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
1A0F
 
 | | CRYSTAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE FROM ESCHERICHIA COLI COMPLEXED WITH GLUTATHIONESULFONIC ACID | | Descriptor: | GLUTATHIONE S-TRANSFERASE, GLUTATHIONE SULFONIC ACID | | Authors: | Nishida, M, Harada, S, Noguchi, S, Inoue, H, Takahashi, K, Satow, Y. | | Deposit date: | 1997-11-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of Escherichia coli glutathione S-transferase complexed with glutathione sulfonate: catalytic roles of Cys10 and His106.
J.Mol.Biol., 281, 1998
|
|
6QUB
 
 | | Truncated beta-galactosidase III from Bifidobacterium bifidum in complex with galactose | | Descriptor: | Beta-galactosidase, CALCIUM ION, beta-D-galactopyranose | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
7FO6
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07H04 from the F2X-Universal Library | | Descriptor: | 1-[(2S)-2-methylmorpholin-4-yl]-2-(thiophen-3-yl)ethan-1-one, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FJY
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04A02 from the F2X-Universal Library | | Descriptor: | 1-[(3-aminophenyl)methyl]piperidin-4-ol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FK4
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04A11 from the F2X-Universal Library | | Descriptor: | (4-methylpiperazin-1-yl)(5-methylthiophen-3-yl)methanone, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
5Z0E
 
 | |
