6HV7
 
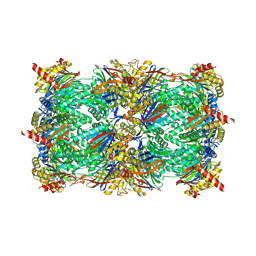 | |
7Q35
 
 | | Crystal structure of the mutant bacteriorhodopsin pressurized with krypton | | Descriptor: | Bacteriorhodopsin, EICOSANE, HEXANE, ... | | Authors: | Melnikov, I, Rulev, M, Astashkin, R, Kovalev, K, Carpentier, P, Gordeliy, V, Popov, A. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-pressure crystallography shows noble gas intervention into protein-lipid interaction and suggests a model for anaesthetic action.
Commun Biol, 5, 2022
|
|
6HW8
 
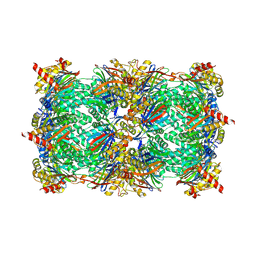 | | Yeast 20S proteasome in complex with 39 | | Descriptor: | (2~{S})-~{N}-[(3~{S},4~{R})-1-cyclohexyl-5-methyl-4,5-bis(oxidanyl)hexan-3-yl]-3-(4-methoxyphenyl)-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
7DBS
 
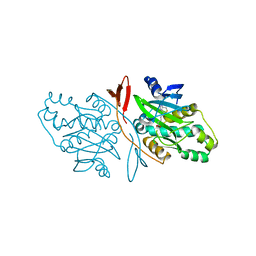 | |
7PXY
 
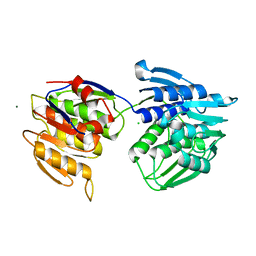 | |
6HM8
 
 | |
7Q37
 
 | | Crystal structure of proton pump MAR rhodopsin pressurized with krypton | | Descriptor: | Bacteriorhodopsin, EICOSANE, HEXANE, ... | | Authors: | Melnikov, I, Rulev, M, Astashkin, R, Kovalev, K, Carpentier, P, Gordeliy, V, Popov, A. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-pressure crystallography shows noble gas intervention into protein-lipid interaction and suggests a model for anaesthetic action.
Commun Biol, 5, 2022
|
|
7Q36
 
 | | Crystal structure of KR2 sodium pump rhodopsin pressurized with krypton | | Descriptor: | EICOSANE, HEXANE, KRYPTON, ... | | Authors: | Melnikov, I, Rulev, M, Astashkin, R, Kovalev, K, Carpentier, P, Gordeliy, V, Popov, A. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-pressure crystallography shows noble gas intervention into protein-lipid interaction and suggests a model for anaesthetic action.
Commun Biol, 5, 2022
|
|
7Q38
 
 | | Crystal structure of the mutant bacteriorhodopsin pressurized with argon | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ARGON, Bacteriorhodopsin, ... | | Authors: | Melnikov, I, Rulev, M, Astashkin, R, Kovalev, K, Carpentier, P, Gordeliy, V, Popov, A. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-pressure crystallography shows noble gas intervention into protein-lipid interaction and suggests a model for anaesthetic action.
Commun Biol, 5, 2022
|
|
1EXK
 
 | | SOLUTION STRUCTURE OF THE CYSTEINE-RICH DOMAIN OF THE ESCHERICHIA COLI CHAPERONE PROTEIN DNAJ. | | Descriptor: | DNAJ PROTEIN, ZINC ION | | Authors: | Martinez-Yamout, M, Legge, G.B, Zhang, O, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-05-03 | | Release date: | 2000-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ.
J.Mol.Biol., 300, 2000
|
|
7PGY
 
 | | Structure of light-adapted AsLOV2 wild type | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Gelfert, R, Weyand, M, Moeglich, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Signal transduction in light-oxygen-voltage receptors lacking the active-site glutamine.
Nat Commun, 13, 2022
|
|
1EAN
 
 | | THE RUNX1 Runt domain at 1.70A resolution: A structural switch and specifically bound chloride ions modulate DNA binding | | Descriptor: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T, Grundstrom, T, Sauer, U.H. | | Deposit date: | 2001-07-13 | | Release date: | 2002-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Runx1 Runt Domain at 1.25 A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
1EHH
 
 | |
1EI2
 
 | | STRUCTURAL BASIS FOR RECOGNITION OF THE RNA MAJOR GROOVE IN THE TAU EXON 10 SPLICING REGULATORY ELEMENT BY AMINOGLYCOSIDE ANTIBIOTICS | | Descriptor: | NEOMYCIN, TAU EXON 10 SRE RNA | | Authors: | Varani, L, Spillantini, M.G, Goedert, M, Varani, G. | | Deposit date: | 2000-02-23 | | Release date: | 2000-03-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the RNA major groove in the tau exon 10 splicing regulatory element by aminoglycoside antibiotics.
Nucleic Acids Res., 28, 2000
|
|
7PGX
 
 | | Structure of dark-adapted AsLOV2 wild type | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Gelfert, R, Weyand, M, Moeglich, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.001 Å) | | Cite: | Signal transduction in light-oxygen-voltage receptors lacking the active-site glutamine.
Nat Commun, 13, 2022
|
|
3CLL
 
 | | Crystal structure of the Spinach Aquaporin SoPIP2;1 S115E mutant | | Descriptor: | Aquaporin | | Authors: | Nyblom, M, Alfredsson, A, Hallgren, K, Hedfalk, K, Neutze, R, Trnroth-Horsefield, S. | | Deposit date: | 2008-03-19 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating.
J.Mol.Biol., 387, 2009
|
|
3CWF
 
 | | Crystal structure of PAS domain of two-component sensor histidine kinase | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alkaline phosphatase synthesis sensor protein phoR | | Authors: | Chang, C, Tesar, C, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-21 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Extracytoplasmic PAS-like domains are common in signal transduction proteins.
J.Bacteriol., 192, 2010
|
|
1EM6
 
 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH GLCNAC AND CP-526,423 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BIS[5-CHLORO-1H-INDOL-2-YL-CARBONYL-AMINOETHYL]-ETHYLENE GLYCOL, LIVER GLYCOGEN PHOSPHORYLASE, ... | | Authors: | Rath, V.L, Ammirati, M, Danley, D.E, Ekstrom, J.L, Hynes, T.R, Olson, T.V, Hoover, D.J. | | Deposit date: | 2000-03-16 | | Release date: | 2000-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human liver glycogen phosphorylase inhibitors bind at a new allosteric site.
Chem.Biol., 7, 2000
|
|
7UFR
 
 | | Cryo-EM Structure of Bl_Man38A at 2.7 A | | Descriptor: | Alpha-mannosidase, ZINC ION | | Authors: | Santos, C.R, Cordeiro, R.L, Domingues, M.N, Borges, A.C, de Farias, M.A, Van Heel, M, Murakami, M.T, Portugal, R.V. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM Structure of Bl_Man38A at 2.7 A
Nat.Chem.Biol., 2022
|
|
6JE8
 
 | | crystal structure of a beta-N-acetylhexosaminidase | | Descriptor: | Beta-N-acetylhexosaminidase, FORMIC ACID, GLYCEROL, ... | | Authors: | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | Deposit date: | 2019-02-04 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
7PGZ
 
 | | Structure of dark-adapted AsLOV2 Q513L | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Gelfert, R, Weyand, M, Moeglich, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Signal transduction in light-oxygen-voltage receptors lacking the active-site glutamine.
Nat Commun, 13, 2022
|
|
7PH0
 
 | | Structure of light-adapted AsLOV2 Q513L | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Gelfert, R, Weyand, M, Moeglich, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.979 Å) | | Cite: | Signal transduction in light-oxygen-voltage receptors lacking the active-site glutamine.
Nat Commun, 13, 2022
|
|
7UFU
 
 | | Cryo-EM Structure of Bl_Man38A nucleophile mutant in complex with mannose at 2.7 A | | Descriptor: | Alpha-mannosidase, ZINC ION, alpha-D-mannopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Domingues, M.N, Borges, A.C, de Farias, M.A, Van Heel, M, Murakami, M.T, Portugal, R.V. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM Structure of Bl_Man38A nucleophile mutant in complex with mannose at 2.7 A
Nat.Chem.Biol., 2022
|
|
7Q0P
 
 | | Structure of the Candida albicans 80S ribosome in complex with anisomycin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
8WKY
 
 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S25 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, N-(2-aminoethyl)-5-(2-{[4-(morpholin-4-yl)pyridin-2-yl]amino}-1,3-thiazol-5-yl)pyridine-3-carboxamide, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
