2JSS
 
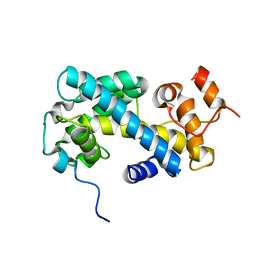 | | NMR structure of chaperone Chz1 complexed with histone H2A.Z-H2B | | Descriptor: | Chimera of Histone H2B.1 and Histone H2A.Z, Uncharacterized protein YER030W | | Authors: | Zhou, Z, Feng, H, Hansen, D.F, Kato, H, Luk, E, Freedberg, D.I, Kay, L.E, Wu, C, Bai, Y. | | Deposit date: | 2007-07-11 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1GQM
 
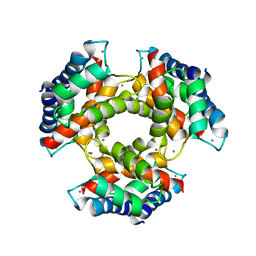 | | The structure of S100A12 in a hexameric form and its proposed role in receptor signalling | | Descriptor: | CALCIUM ION, CALGRANULIN C | | Authors: | Moroz, O.V, Antson, A.A, Dodson, E.G, Burrel, H.J, Grist, S.J, Lloyd, R.M, Maitland, N.J, Dodson, G.G, Wilson, K.S, Lukanidin, E, Bronstein, I.B. | | Deposit date: | 2001-11-26 | | Release date: | 2002-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of S100A12 in a Hexameric Form and its Proposed Role in Receptor Signalling
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8QU9
 
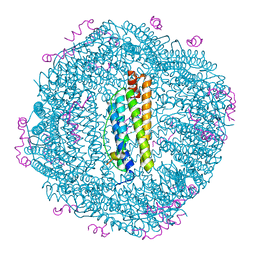 | | Structure of the NCOA4 (Nuclear Receptor Coactivator 4)-FTH1 (H-Ferritin) complex | | Descriptor: | FE (III) ION, Ferritin heavy chain, NCOA4 (Nuclear Receptor Coactivator 4) | | Authors: | Hoelzgen, F, Klukin, E, Zalk, R, Shahar, A, Cohen-Schwartz, S, Frank, G.A. | | Deposit date: | 2023-10-15 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for the intracellular regulation of ferritin degradation.
Nat Commun, 15, 2024
|
|
1E8A
 
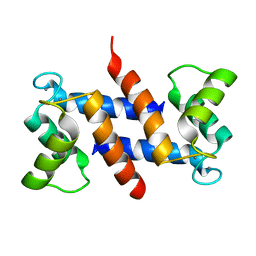 | | The three-dimensional structure of human S100A12 | | Descriptor: | CALCIUM ION, S100A12 | | Authors: | Moroz, O.V, Antson, A.A, Murshudov, G.N, Maitland, N.J, Dodson, G.G, Wilson, K.S, Skibshoj, I, Lukanidin, E.M, Bronstein, I.B. | | Deposit date: | 2000-09-18 | | Release date: | 2001-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Three-Dimensional Structure of Human S100A12
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1AUL
 
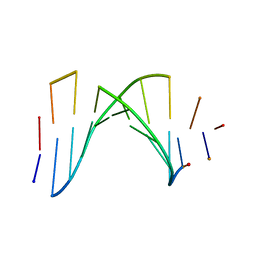 | | SOLUTION STRUCTURE OF A HIGHLY STABLE DNA DUPLEX CONJUGATED TO A MINOR GROOVE BINDER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*TP*AP*AP*TP*CP*A)-3'), DNA (5'-D(P*THXP*GP*AP*TP*TP*AP*TP*CP*TP*G)-3') | | Authors: | Kumar, S, Reed, M.W, Gamper Junior, H.B, Gorn, V.V, Lukhtanov, E.A, Foti, M, West, J, Meyer Junior, R.B, Schweitzer, B.I. | | Deposit date: | 1997-08-29 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a highly stable DNA duplex conjugated to a minor groove binder.
Nucleic Acids Res., 26, 1998
|
|
1ODB
 
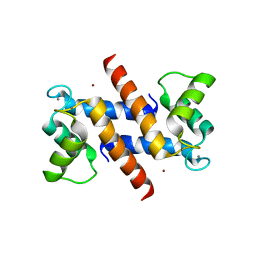 | | THE CRYSTAL STRUCTURE OF HUMAN S100A12 - COPPER COMPLEX | | Descriptor: | CALCIUM ION, CALGRANULIN C, COPPER (II) ION | | Authors: | Moroz, O.V, Antson, A.A, Grist, S.J, Maitland, N.J, Dodson, G.G, Wilson, K.S, Lukanidin, E.M, Bronstein, I.B. | | Deposit date: | 2003-02-15 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the Human S100A12-Copper Complex: Implications for Host-Parasite Defence
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5NZ4
 
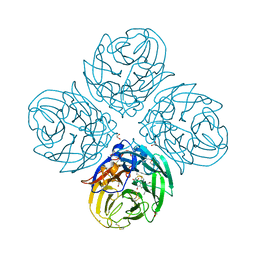 | | Complex of I223V mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J, Karlukova, E. | | Deposit date: | 2017-05-12 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
