1S2T
 
 | | Crystal Structure Of Apo Phosphoenolpyruvate Mutase | | Descriptor: | Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2U
 
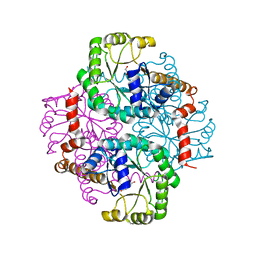 | | Crystal structure of the D58A phosphoenolpyruvate mutase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
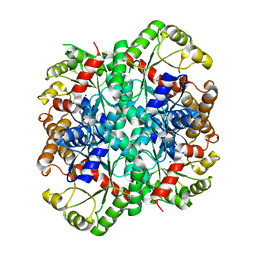
1S2W
 
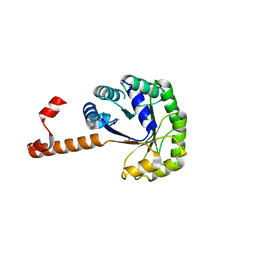 | | Crystal structure of phosphoenolpyruvate mutase in high ionic strength | | Descriptor: | Phosphoenolpyruvate phosphomutase, SULFATE ION | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2V
 
 | | Crystal structure of phosphoenolpyruvate mutase complexed with Mg(II) | | Descriptor: | MAGNESIUM ION, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
3T8S
 
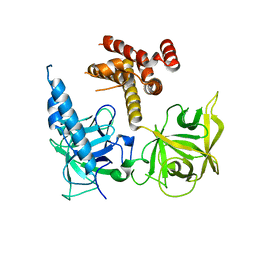 | | Apo and InsP3-bound Crystal Structures of the Ligand-Binding Domain of an InsP3 Receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Lin, C, Baek, K, Lu, Z. | | Deposit date: | 2011-08-01 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Apo and InsP(3)-bound crystal structures of the ligand-binding domain of an InsP(3) receptor.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3UJT
 
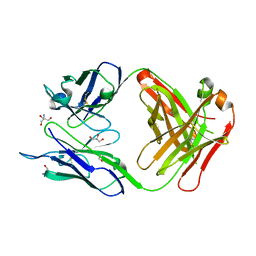 | | Structure of the Fab fragment of Ab-52, an antibody that binds the O-antigen of Francisella tularensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ab-52 heavy chain, Ab-52 light chain, ... | | Authors: | Rynkiewicz, M.J, Lu, Z, Hui, J.H, Sharon, J, Seaton, B.A. | | Deposit date: | 2011-11-08 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of a Protective Epitope of the Francisella tularensis O-Polysaccharide.
Biochemistry, 51, 2012
|
|
6BNS
 
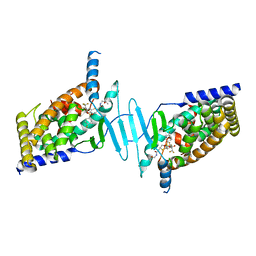 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN BOUND TETHERED WITH SRC co-activator peptide and Compound 25a AKA BICYCLIC HEXAFLUOROISOPROPYL 2 ALCOHOL SULFONAMIDES | | Descriptor: | 2-[(2S)-4-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)-3,4-dihydro-2H-1,4-benzothiazin-2-yl]-N-(2-hydroxy-2-methylpropyl)acetamide, Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1 Chimera | | Authors: | DHAR, T.G, GONG, H, WEINSTEIN, D.S, LU, Z, DUAN, J.J.W, STACHURA, S, HAQUE, L, KARMAKAR, A, HEMAGIRI, H, RAUT, D.K, GUPTA, A.K, KHAN, J.A, SACK, J.S, CAMAC, D.M, PUDZIANOWSKI, A.A, WU, D.R, YARDE, M, SHEN, D.R, BOROWSKI, V, XIE, J.H, SUN, H, ARIENZO, C.D, DABROS, M, GALELLA, M.A, WANG, F, WEIGELT, C.A, ZHAO, Q, FOSTER, W, SOMERVILLE, J.E, SALTER-CID, L.M, BARRISH, J.C, CARTER, P.H. | | Deposit date: | 2017-11-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Identification of bicyclic hexafluoroisopropyl alcohol sulfonamides as retinoic acid receptor-related orphan receptor gamma (ROR gamma /RORc) inverse agonists. Employing structure-based drug design to improve pregnane X receptor (PXR) selectivity.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5WIE
 
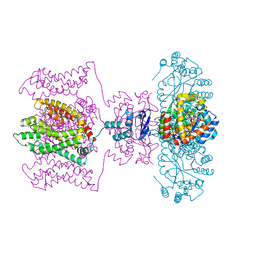 | | Crystal structure of a Kv1.2-2.1 chimera K+ channel V406W mutant in an inactivated state | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Pau, V, Zhou, Y, Ramu, Y, Xu, Y, Lu, Z. | | Deposit date: | 2017-07-19 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of an inactivated mutant mammalian voltage-gated K(+) channel.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7X2P
 
 | |
7ZOI
 
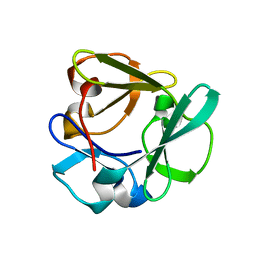 | | Carbohydrate binding domain CBM92-A from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 | | Descriptor: | Glycoside hydrolase family 18 | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOO
 
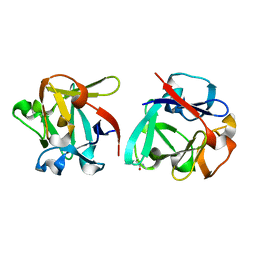 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with gentiobiose | | Descriptor: | Glycoside hydrolase family 18, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZON
 
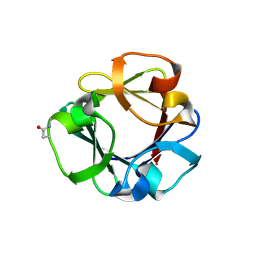 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with glucose | | Descriptor: | Glycoside hydrolase family 18, PENTAETHYLENE GLYCOL, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOH
 
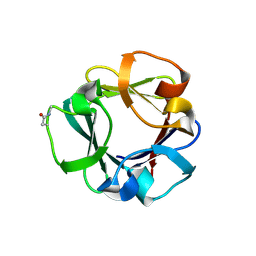 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 | | Descriptor: | Glycoside hydrolase family 18 | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOP
 
 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with sophorose. | | Descriptor: | Glycoside hydrolase family 18, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
