7XX4
 
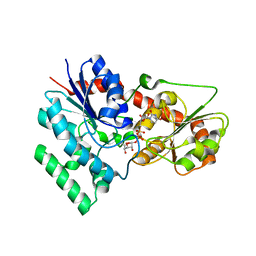 | | designed glycosyltransferase | | Descriptor: | Oleandomycin glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Lu, M, Wu, X. | | Deposit date: | 2022-05-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Design of a chimeric glycosyltransferase OleD for the site-specific O-monoglycosylation of 3-hydroxypyridine in nosiheptide.
Microb Biotechnol, 16, 2023
|
|
6VS0
 
 | | protein B | | Descriptor: | CHLORAMPHENICOL, Multidrug transporter MdfA, PRASEODYMIUM ION | | Authors: | Lu, M, Lu, M.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of a redesigned multidrug transporter from the Major Facilitator Superfamily.
Sci Rep, 10, 2020
|
|
6VS2
 
 | | protein D | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Multidrug transporter MdfA, PRASEODYMIUM ION | | Authors: | Lu, M, Lu, M.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanism of a redesigned multidrug transporter from the Major Facilitator Superfamily.
Sci Rep, 10, 2020
|
|
6VS1
 
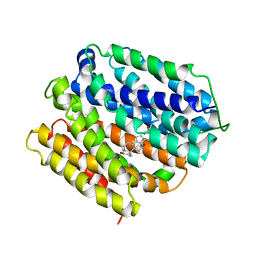 | | protein C | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Multidrug transporter MdfA, PRASEODYMIUM ION | | Authors: | Lu, M, Lu, M.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanism of a redesigned multidrug transporter from the Major Facilitator Superfamily.
Sci Rep, 10, 2020
|
|
6VRZ
 
 | | protein A | | Descriptor: | CHLORAMPHENICOL, Multidrug transporter MdfA, PRASEODYMIUM ION | | Authors: | Lu, M, Lu, M.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of a redesigned multidrug transporter from the Major Facilitator Superfamily.
Sci Rep, 10, 2020
|
|
4XWW
 
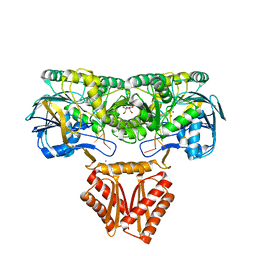 | | Crystal structure of RNase J complexed with RNA | | Descriptor: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | Deposit date: | 2015-01-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
6X63
 
 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WAP
 
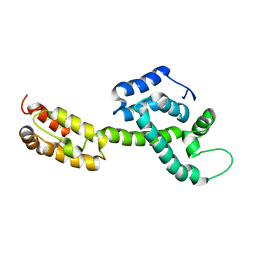 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4XWT
 
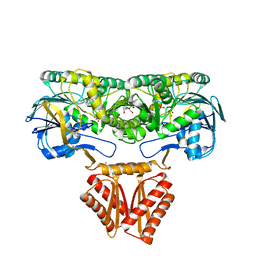 | | Crystal structure of RNase J complexed with UMP | | Descriptor: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | Deposit date: | 2015-01-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
6OOM
 
 | | PROTEIN A | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Multidrug transporter MdfA, PRASEODYMIUM ION | | Authors: | Lu, M. | | Deposit date: | 2019-04-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an engineered multidrug transporter MdfA reveals the molecular basis for substrate recognition.
Commun Biol, 2, 2019
|
|
6OOP
 
 | | protein B | | Descriptor: | 1,1'-dimethyl-4,4'-bipyridin-1-ium, Multidrug transporter MdfA, PRASEODYMIUM ION | | Authors: | Lu, M. | | Deposit date: | 2019-04-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an engineered multidrug transporter MdfA reveals the molecular basis for substrate recognition.
Commun Biol, 2, 2019
|
|
2QFI
 
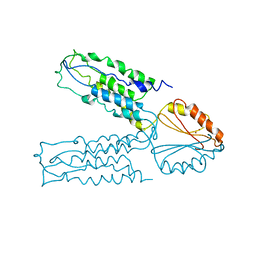 | | Structure of the zinc transporter YiiP | | Descriptor: | Ferrous-iron efflux pump fieF, ZINC ION | | Authors: | Lu, M. | | Deposit date: | 2007-06-27 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the zinc transporter YiiP.
Science, 317, 2007
|
|
3H90
 
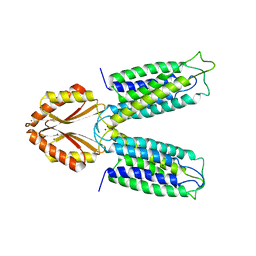 | |
3HFH
 
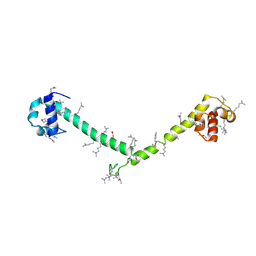 | | Crystal structure of tandem FF domains | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Lu, M, Yang, J, Ren, Z, Subir, S, Bedford, M.T, Jacobson, R.H, McMurray, J.S, Chen, X. | | Deposit date: | 2009-05-11 | | Release date: | 2009-08-18 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal Structure of the Three Tandem FF Domains of the Transcription Elongation Regulator CA150.
J.Mol.Biol., 393, 2009
|
|
6OOQ
 
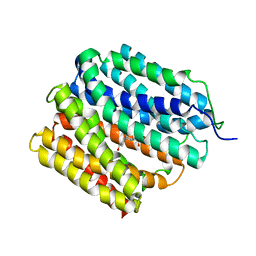 | | protein C | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Multidrug transporter MdfA | | Authors: | Lu, M. | | Deposit date: | 2019-04-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an engineered multidrug transporter MdfA reveals the molecular basis for substrate recognition.
Commun Biol, 2, 2019
|
|
1DFU
 
 | |
4OI6
 
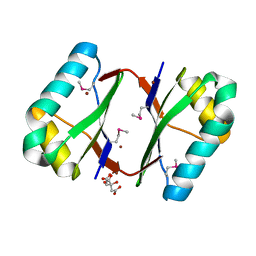 | | Crystal structure analysis of nickel-bound form SCO4226 from Streptomyces coelicolor A3(2) | | Descriptor: | CITRIC ACID, NICKEL (II) ION, Nickel responsive protein | | Authors: | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-01-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
4OI3
 
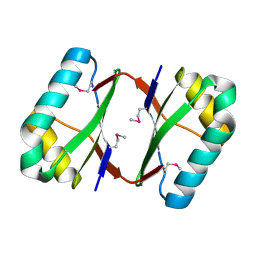 | | Crystal structure analysis of SCO4226 from Streptomyces coelicolor A3(2) | | Descriptor: | Nickel responsive protein | | Authors: | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-01-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
7TA8
 
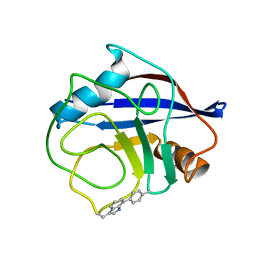 | | NMR structure of crosslinked cyclophilin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Lu, M, Toptygin, D, Xiang, Y, Shi, Y, Schwieters, C.D, Lipinski, E.C, Ahn, J, Byeon, I.-J.L, Gronenborn, A.M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The Magic of Linking Rings: Discovery of a Unique Photoinduced Fluorescent Protein Crosslink.
J.Am.Chem.Soc., 144, 2022
|
|
8H4H
 
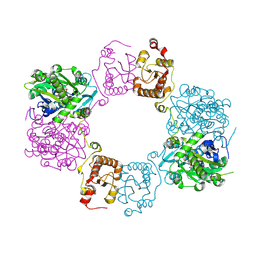 | |
2FUN
 
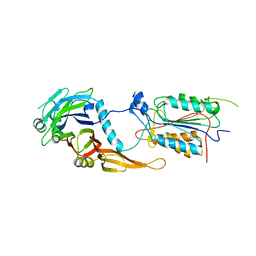 | | alternative p35-caspase-8 complex | | Descriptor: | Early 35 kDa protein, caspase-8 | | Authors: | Lu, M, Min, T, Eliezer, D, Wu, H. | | Deposit date: | 2006-01-27 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Native chemical ligation in covalent caspase inhibition by p35.
Chem.Biol., 13, 2006
|
|
1CE9
 
 | | HELIX CAPPING IN THE GCN4 LEUCINE ZIPPER | | Descriptor: | PROTEIN (GCN4-PMSE) | | Authors: | Lu, M, Shu, W, Ji, H, Spek, E, Wang, L.-Y, Kallenbach, N.R. | | Deposit date: | 1999-03-18 | | Release date: | 1999-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Helix capping in the GCN4 leucine zipper.
J.Mol.Biol., 288, 1999
|
|
5C6O
 
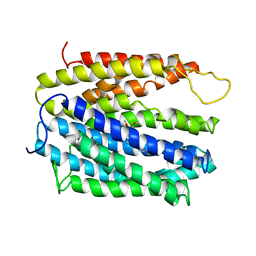 | | protein B | | Descriptor: | (2S)-2-(3,4-dimethoxyphenyl)-5-{[2-(3,4-dimethoxyphenyl)ethyl](methyl)amino}-2-(propan-2-yl)pentanenitrile, BH2163 protein | | Authors: | Lu, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the blockade of MATE multidrug efflux pumps.
Nat Commun, 6, 2015
|
|
5ULD
 
 | |
5UL9
 
 | |
