6YVU
 
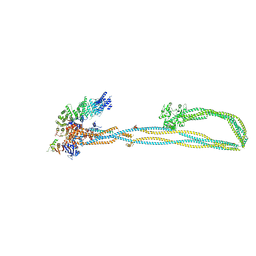 | | Condensin complex from S.cerevisiae ATP-free apo non-engaged state | | Descriptor: | Condensin complex subunit 1,Condensin complex subunit 1,Ycs4, Condensin complex subunit 2,Condensin complex subunit 2,Brn1, Structural maintenance of chromosomes protein 2,Structural maintenance of chromosomes protein 2,Smc2, ... | | Authors: | Lee, B.-G, Cawood, C, Gutierrez-Escribano, P, Nakane, T, Merkel, F, Hassler, M, Aragon, L, Haering, C.H, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4MJD
 
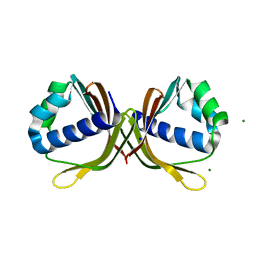 | | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747 | | Descriptor: | Ketosteroid isomerase fold protein Hmuk_0747, MAGNESIUM ION, SODIUM ION | | Authors: | Chang, C, Holowicki, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-03 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747
To be Published
|
|
8AHL
 
 | |
8AFH
 
 | |
8AJB
 
 | |
7PB9
 
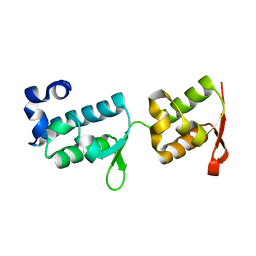 | | Crystal structure of tandem WH domains of Vps25 from Odinarchaeota | | Descriptor: | Tandem WH domains of Vps25 | | Authors: | Salzer, R, Bellini, D, Papatziamou, D, Robinson, N.P, Lowe, J. | | Deposit date: | 2021-08-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asgard archaea shed light on the evolutionary origins of the eukaryotic ubiquitin-ESCRT machinery.
Nat Commun, 13, 2022
|
|
8ONW
 
 | |
7P6U
 
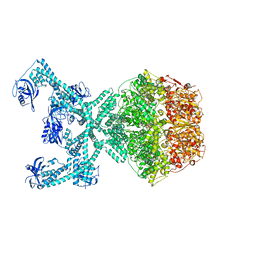 | | Lon protease from Thermus Thermophilus | | Descriptor: | (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK), Lon protease, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Coscia, F, Lowe, J. | | Deposit date: | 2021-07-18 | | Release date: | 2021-10-27 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the full-length Lon protease from Thermus thermophilus.
Febs Lett., 595, 2021
|
|
8AFM
 
 | |
4ML9
 
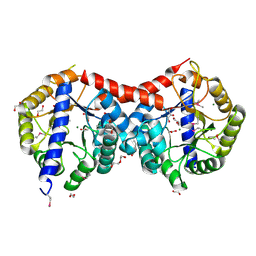 | | Crystal Structure of Uncharacterized TIM Barrel Protein with the Conserved Phosphate Binding Site fromSebaldella termitidis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Crystal Structure of Uncharacterized TIM Barrel Protein with the Conserved Phosphate Binding Site fromSebaldella termitidis
To be Published
|
|
8BH1
 
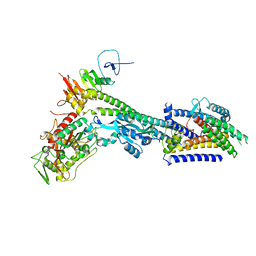 | | Core divisome complex FtsWIQBL from Pseudomonas aeruginosa | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | Authors: | Kaeshammer, L, van den Ent, F, Jeffery, M, Lowe, J. | | Deposit date: | 2022-10-28 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the bacterial divisome core complex and antibiotic target FtsWIQBL.
Nat Microbiol, 8, 2023
|
|
1E4F
 
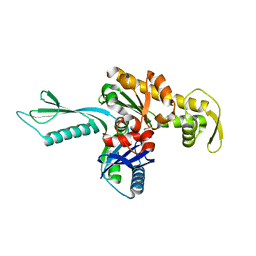 | | FtsA (apo form) from Thermotoga maritima | | Descriptor: | CELL DIVISION PROTEIN FTSA | | Authors: | van den Ent, F, Lowe, J. | | Deposit date: | 2000-07-03 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Cell Division Protein Ftsa from Thermotoga Maritima
Embo J., 19, 2000
|
|
1E4G
 
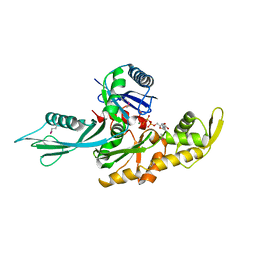 | | FtsA (ATP-bound form) from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN FTSA, MAGNESIUM ION | | Authors: | van den Ent, F, Lowe, J. | | Deposit date: | 2000-07-03 | | Release date: | 2000-10-18 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Cell Division Protein Ftsa from Thermotoga Maritima
Embo J., 19, 2000
|
|
6T8G
 
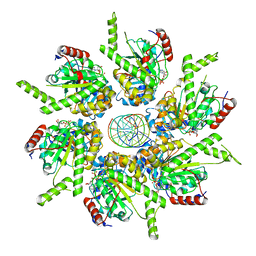 | | Stalled FtsK motor domain bound to dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA translocase FtsK, dsDNA substrate | | Authors: | Jean, N.L, Lowe, J. | | Deposit date: | 2019-10-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | FtsK in motion reveals its mechanism for double-stranded DNA translocation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6H2X
 
 | | MukB coiled-coil elbow from E. coli | | Descriptor: | Chromosome partition protein MukB,Chromosome partition protein MukB | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2018-07-16 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A folded conformation of MukBEF and cohesin.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7NYX
 
 | |
7NYW
 
 | | Cryo-EM structure of the MukBEF-MatP-DNA head module | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NYZ
 
 | |
7NZ0
 
 | |
6T8B
 
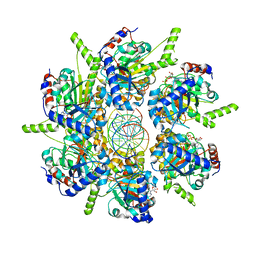 | | FtsK motor domain with dsDNA, translocating state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA translocase FtsK, MAGNESIUM ION, ... | | Authors: | Jean, N.L, Lowe, J. | | Deposit date: | 2019-10-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | FtsK in motion reveals its mechanism for double-stranded DNA translocation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7QUQ
 
 | | BtubA(R284G,K286D,F287G):BtubB bacterial tubulin M-loop mutant forming a single protofilament (Prosthecobacter dejongeii) | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Tubulin beta, Tubulin domain-containing protein | | Authors: | Wagstaff, J, Planelles-Herrero, V.J, Derivery, E, Lowe, J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Diverse cytomotive actins and tubulins share a polymerization switch mechanism conferring robust dynamics.
Sci Adv, 9, 2023
|
|
7QUP
 
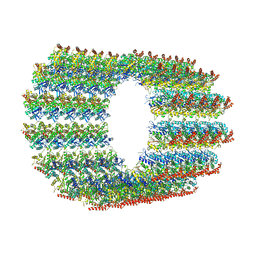 | | D. melanogaster 13-protofilament microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wagstaff, J, Planelles-Herrero, V.J, Derivery, E, Lowe, J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Diverse cytomotive actins and tubulins share a polymerization switch mechanism conferring robust dynamics.
Sci Adv, 9, 2023
|
|
7NYY
 
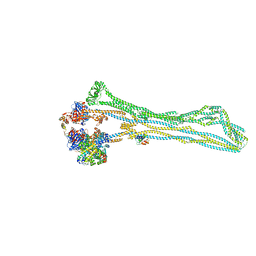 | | Cryo-EM structure of the MukBEF monomer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NZ4
 
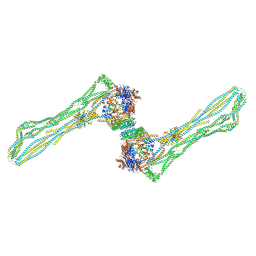 | | Cryo-EM structure of the MukBEF dimer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NZ2
 
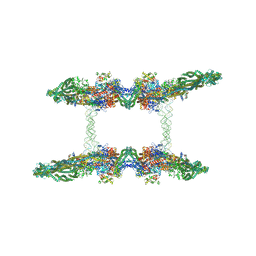 | | Cryo-EM structure of the MukBEF-MatP-DNA tetrad | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
