7SNX
 
 | | 1.70A Resolution Structure of NanoBiT Complementation Reporter Complex of LgBit and SmBiT Subunits | | Descriptor: | GLYCEROL, Oplophorus-luciferin 2-monooxygenase catalytic subunit, Oplophorus-luciferin 2-monooxygenase catalytic subunit: C-terminal Peptide (11-mer) | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70A Resolution Structure of NanoBiT Complementation Reporter Complex of LgBit and SmBiT Subunits
To be published
|
|
3MHZ
 
 | | 1.7A structure of 2-fluorohistidine labeled Protective Antigen | | Descriptor: | CALCIUM ION, Protective antigen, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Wimalasena, D.S, Janowiak, B.E, Miyagi, M, Sun, J, Hajduch, J, Pooput, C, Kirk, K.L, Bann, J.G. | | Deposit date: | 2010-04-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence that histidine protonation of receptor-bound anthrax protective antigen is a trigger for pore formation.
Biochemistry, 49, 2010
|
|
3R2O
 
 | | 1.95 A resolution structure of As-Isolated FtnA from Pseudomonas aeruginosa (pH 6.0) | | Descriptor: | Bacterioferritin, SODIUM ION, SULFATE ION | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
8UA5
 
 | | Crystal Structure of infected cell protein 0 (ICP0) from herpes simplex virus 1 (A636-Q776) | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Lovell, S, Kashipathy, M, Battaile, K.P, Cooper, A, Davido, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | HSV-1 ICP0 dimer domain adopts a novel beta-barrel fold.
Proteins, 92, 2024
|
|
3R2M
 
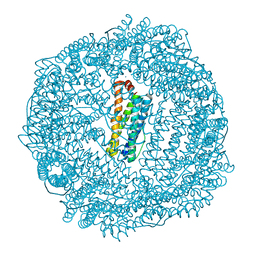 | | 1.8A resolution structure of Doubly Soaked FtnA from Pseudomonas aeruginosa (pH 7.5) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
3R2H
 
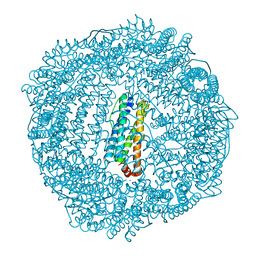 | | 1.7 A resolution structure of As-Isolated FtnA from Pseudomonas aeruginosa (pH 10.5) | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Bacterioferritin, SODIUM ION | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
3R2K
 
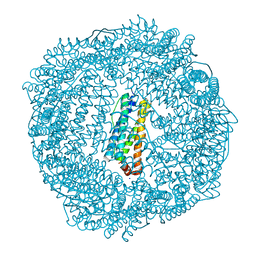 | | 1.55A resolution structure of As-Isolated FtnA from Pseudomonas aeruginosa (pH 7.5) | | Descriptor: | Bacterioferritin, SODIUM ION | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
8UA2
 
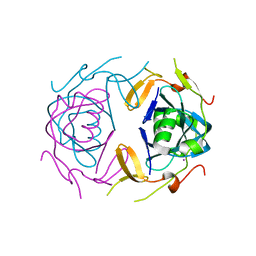 | | Crystal Structure of infected cell protein 0 (ICP0) from herpes simplex virus 1 (proteolyzed fragment) | | Descriptor: | IODIDE ION, RL2 | | Authors: | Lovell, S, Kashipathy, M, Battaile, K.P, Cooper, A, Davido, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | HSV-1 ICP0 dimer domain adopts a novel beta-barrel fold.
Proteins, 92, 2024
|
|
3R2L
 
 | | 1.85A resolution structure of Iron Soaked FtnA from Pseudomonas aeruginosa (pH 7.5) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
3R2S
 
 | | 2.1A resolution structure of Doubly Soaked FtnA from Pseudomonas aeruginosa (pH 6.0) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION, ... | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
3R9J
 
 | | 4.3A resolution structure of a MinD-MinE(I24N) protein complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
3R2R
 
 | | 1.65A resolution structure of Iron Soaked FtnA from Pseudomonas aeruginosa (pH 6.0) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION, ... | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
8F45
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a phenyl dimethyl sulfane inhibitor (cyclopropyl ketoamide warhead) | | Descriptor: | (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(2~{S},3~{S})-3-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
7SNS
 
 | | 1.55A Resolution Structure of NanoLuc Luciferase | | Descriptor: | ACETATE ION, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55A Resolution Structure of NanoLuc Luciferase
To be published
|
|
9DN4
 
 | | Crystal structure of a SARS-CoV-2 20-mer RNA in complex with FAB BL3-6S97N | | Descriptor: | CHLORIDE ION, FAB BL3-6S97N HEAVY CHAIN, FAB BL3-6S97N LIGHT CHAIN, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Hegde, S, Wang, J. | | Deposit date: | 2024-09-16 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a SARS-CoV-2 20-mer RNA in complex with FAB BL3-6S97N
To be published
|
|
8EJX
 
 | |
8W1D
 
 | | CRYSTAL STRUCTURE OF DPS-LIKE PROTEIN PA4880 FROM PSEUDOMONAS AERUGINOSA (DIMERIC FORM) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION | | Authors: | Lovell, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
7SNW
 
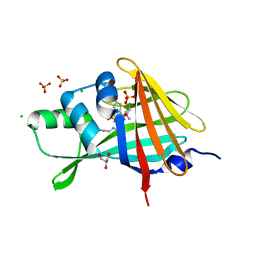 | | 1.80A Resolution Structure of NanoLuc Luciferase with Bound Inhibitor PC 16026576 | | Descriptor: | 2-(methoxycarbonyl)thiophene-3-sulfonic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80A Resolution Structure of NanoLuc Luciferase with Bound Inhibitor PC 16026576
To be published
|
|
9BTS
 
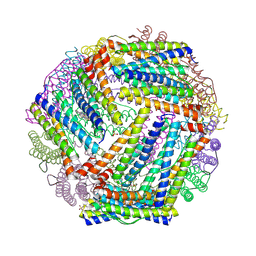 | | Crystal structure of the bacterioferritin (Bfr) and ferritin (Ftn) heterooligomer complex from Acinetobacter baumannii | | Descriptor: | Bacterioferritin (Bfr), Ferritin (Ftn), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of Acinetobacter baumannii bacterioferritin reveals a heteropolymer of bacterioferritin and ferritin subunits.
Sci Rep, 14, 2024
|
|
7T40
 
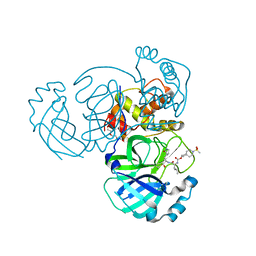 | | Structure of MERS 3CL protease in complex with inhibitor 10c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T49
 
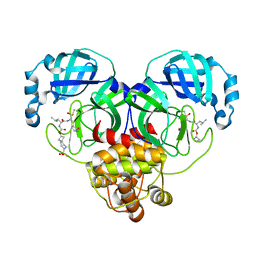 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 10c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T45
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 7c | | Descriptor: | (1S,2S)-2-{[N-({[7-(tert-butoxycarbonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Kankanamalage, A.C.G, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
3R9I
 
 | | 2.6A resolution structure of MinD complexed with MinE (12-31) peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
7TQ7
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 13c | | Descriptor: | N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucinamide, Orf1a protein, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ4
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 6c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-chlorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
