4MPO
 
 | | 1.90 A resolution structure of CT771 from Chlamydia trachomatis Bound to Hydrolyzed Ap4A Products | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, CT771, ... | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2013-09-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chlamydia trachomatis CT771 (nudH) Is an Asymmetric Ap4A Hydrolase.
Biochemistry, 53, 2014
|
|
5WKQ
 
 | | 2.10 A resolution structure of IpaB (residues 74-242) from Shigella flexneri | | Descriptor: | Invasin IpaB | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Picking, W.L, Picking, W.D. | | Deposit date: | 2017-07-25 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Using disruptive insertional mutagenesis to identify the in situ structure-function landscape of the Shigella translocator protein IpaB.
Protein Sci., 27, 2018
|
|
6XG7
 
 | | 1.3 A Resolution Structure of the of the NHL Repeat Region of D. melanogaster Thin | | Descriptor: | SULFATE ION, TETRAETHYLENE GLYCOL, Thin, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Bawa, S, Geisbrecht, E.R. | | Deposit date: | 2020-06-17 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Costameric integrin and sarcoglycan protein levels are altered in a Drosophila model for Limb-girdle muscular dystrophy type 2H.
Mol.Biol.Cell, 32, 2021
|
|
4DM0
 
 | | TN5 transposase: 20MER OUTSIDE END 2 MN complex | | Descriptor: | 1,2-ETHANEDIOL, DNA NON-TRANSFERRED STRAND, DNA TRANSFERRED STRAND, ... | | Authors: | Klenchin, V.A, Lovell, S, Goryshin, I.Y, Reznikoff, W.R, Rayment, I. | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two-metal active site binding of a TN5 transposase synaptic complex
Nat.Struct.Biol., 9, 2002
|
|
7TL7
 
 | | 1.90A resolution structure of independent phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Sa-D2) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, IMIDAZOLE, SODIUM ION, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dranchak, P, Queme, B, Aitha, M, van Neer, R.H.P, Kimura, H, Katho, T, Suga, H, Inglese, J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serum-Stable and Selective Backbone-N-Methylated Cyclic Peptides That Inhibit Prokaryotic Glycolytic Mutases.
Acs Chem.Biol., 17, 2022
|
|
7TL8
 
 | | 1.95A resolution structure of independent phosphoglycerate mutase from S. aureus in complex with a macrocyclic peptide inhibitor (Sa-D3) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, MANGANESE (II) ION, Peptide Sa-D3 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dranchak, P, Queme, B, Aitha, M, van Neer, R.H.P, Kimura, H, Katho, T, Suga, H, Inglese, J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Serum-Stable and Selective Backbone-N-Methylated Cyclic Peptides That Inhibit Prokaryotic Glycolytic Mutases.
Acs Chem.Biol., 17, 2022
|
|
1PZ4
 
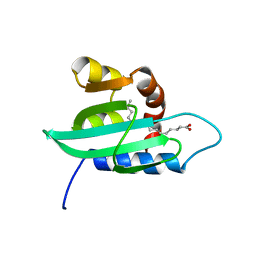 | | The structural determination of an insect (mosquito) Sterol Carrier Protein-2 with a ligand bound C16 Fatty Acid at 1.35 A resolution | | Descriptor: | PALMITIC ACID, sterol carrier protein 2 | | Authors: | Dyer, D.H, Lovell, S, Thoden, J.B, Holden, H.M, Rayment, I, Lan, Q. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Structural Determination of an Insect Sterol Carrier Protein-2 with a Ligand-bound C16 Fatty Acid at 1.35A Resolution
J.Biol.Chem., 278, 2003
|
|
8CZU
 
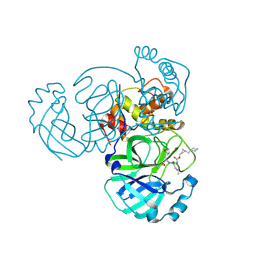 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 16d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{S})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZT
 
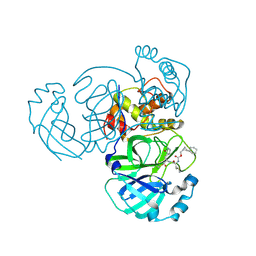 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZV
 
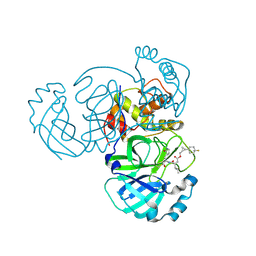 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZX
 
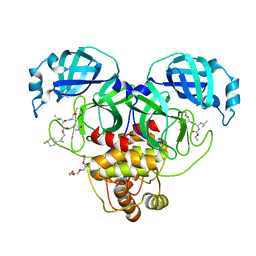 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, TETRAETHYLENE GLYCOL, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, ... | | Authors: | Machen, A.J, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZW
 
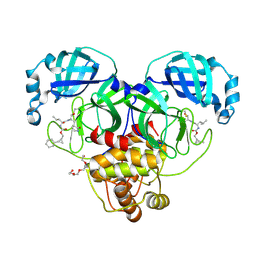 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, TETRAETHYLENE GLYCOL, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, ... | | Authors: | Machen, A.J, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
1P7J
 
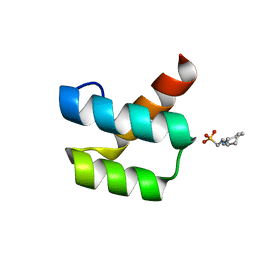 | | Crystal structure of engrailed homeodomain mutant K52E | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
1MM8
 
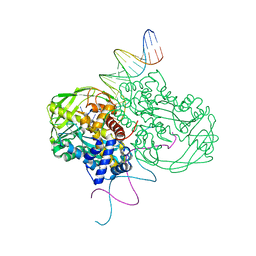 | | Crystal structure of Tn5 Transposase complexed with ME DNA | | Descriptor: | MANGANESE (II) ION, ME DNA non-transferred strand, ME DNA transferred strand, ... | | Authors: | Steiniger-White, M, Bhasin, A, Lovell, S, Rayment, I, Reznikoff, W.S. | | Deposit date: | 2002-09-03 | | Release date: | 2002-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for "unseen" Transposase--DNA contacts
J.Mol.Biol., 322, 2002
|
|
1P7I
 
 | | CRYSTAL STRUCTURE OF ENGRAILED HOMEODOMAIN MUTANT K52A | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
8E6D
 
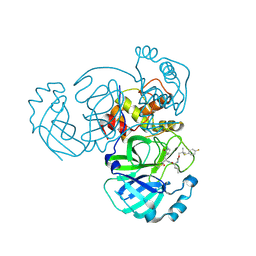 | | Crystal structure of MERS 3CL protease in complex with a p-fluorophenyl dimethyl sulfane inhibitor | | Descriptor: | (1R,2S)-2-{[N-({2-[(4-fluorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
5IQW
 
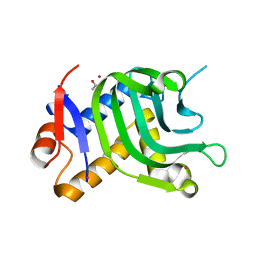 | | 1.95A resolution structure of Apo HasAp (R33A) from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, CADMIUM ION, Heme acquisition protein HasAp | | Authors: | Kumar, R, Lovell, S, Battaile, K.P, Yao, H, Rivera, M. | | Deposit date: | 2016-03-11 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Replacing Arginine 33 for Alanine in the Hemophore HasA from Pseudomonas aeruginosa Causes Closure of the H32 Loop in the Apo-Protein.
Biochemistry, 55, 2016
|
|
5IQX
 
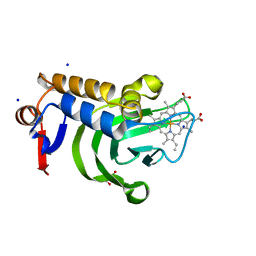 | | 1.05A resolution structure of Holo HasAp (R33A) from Pseudomonas aeruginosa | | Descriptor: | D-MALATE, Heme acquisition protein HasAp, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kumar, R, Lovell, S, Battaile, K.P, Yao, H, Rivera, M. | | Deposit date: | 2016-03-11 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Replacing Arginine 33 for Alanine in the Hemophore HasA from Pseudomonas aeruginosa Causes Closure of the H32 Loop in the Apo-Protein.
Biochemistry, 55, 2016
|
|
8E6C
 
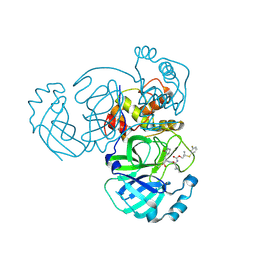 | | Crystal structure of MERS 3CL protease in complex with a m-fluorophenyl dimethyl sulfane inhibitor | | Descriptor: | Orf1a protein, [2-(3-fluorophenyl)sulfanyl-2-methyl-propyl] ~{N}-[(2~{S})-1-[[3-[(3~{S})-2-$l^{3}-oxidanylidenepyrrolidin-3-yl]-1-$l^{1}-oxidanylsulfonyl-1-oxidanyl-propan-2-yl]-$l^{2}-azanyl]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E6B
 
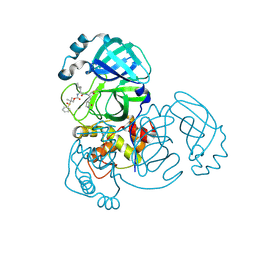 | | Crystal structure of MERS 3CL protease in complex with a dimethyl sulfinyl benzene inhibitor | | Descriptor: | (2~{S})-2-[[(2~{S})-4-methyl-2-[[2-methyl-2-[oxidanyl(phenyl)-$l^{3}-sulfanyl]propoxy]carbonylamino]pentanoyl]amino]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propane-1-sulfonic acid, N~2~-(ethoxycarbonyl)-N-{(1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E6E
 
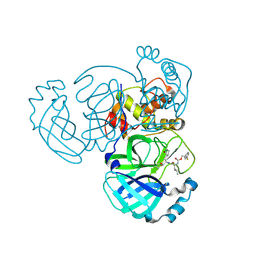 | | Crystal structure of MERS 3CL protease in complex with a phenyl sulfane inhibitor | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[2-(phenylsulfanyl)ethoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid, 2-phenylsulfanylethyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, Orf1a protein | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Madden, T.K, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
6DE6
 
 | |
6CR0
 
 | | 1.55 A resolution structure of (S)-6-hydroxynicotine oxidase from Shinella HZN7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (S)-6-hydroxynicotine oxidase, ACETATE ION, ... | | Authors: | Deay III, D, Lovell, S, Battaile, K.P, Petillo, P, Richter, M. | | Deposit date: | 2018-03-16 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Improving the kinetic parameters of nicotine oxidizing enzymes by homologous structure comparison and rational design
Arch.Biochem.Biophys., 2022
|
|
4JER
 
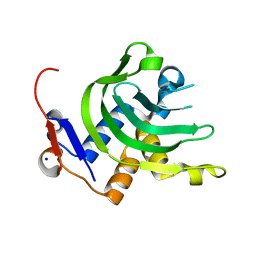 | | 1.1A resolution Apo structure of the hemophore HasA from Yersinia pestis (Tetragonal Form) | | Descriptor: | Hemophore HasA, SODIUM ION | | Authors: | Kumar, R, Lovell, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Hemophore HasA from Yersinia pestis (HasAyp) Coordinates Hemin with a Single Residue, Tyr75, and with Minimal Conformational Change.
Biochemistry, 52, 2013
|
|
4JES
 
 | | 1.6A resolution Apo structure of the hemophore HasA from Yersinia pestis (Hexagonal Form) | | Descriptor: | HEXAETHYLENE GLYCOL, Hemophore HasA, MALONATE ION, ... | | Authors: | Kumar, R, Lovell, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Hemophore HasA from Yersinia pestis (HasAyp) Coordinates Hemin with a Single Residue, Tyr75, and with Minimal Conformational Change.
Biochemistry, 52, 2013
|
|
