1Z5F
 
 | | Solution Structure of the Cytotoxic RC-RNase 3 with a Pyroglutamate Residue at the N-terminus | | Descriptor: | RC-RNase 3 | | Authors: | Lou, Y.C, Huang, Y.C, Pan, Y.R, Chen, C, Liao, Y.D. | | Deposit date: | 2005-03-18 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Roles of N-terminal pyroglutamate in maintaining structural integrity and pKa values of catalytic histidine residues in bullfrog ribonuclease 3
J.Mol.Biol., 355, 2006
|
|
6JK2
 
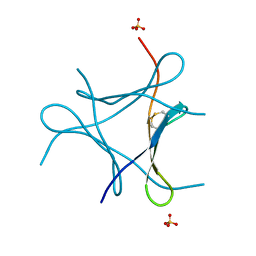 | | Crystal structure of a mini fungal lectin, PhoSL | | Descriptor: | Lectin, SULFATE ION | | Authors: | Lou, Y.C, Chou, C.C, Yeh, H.H, Chien, C.Y, Sushant, S, Chen, C, Hsu, C.H. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structural insights into the role of N-terminal integrity in PhoSL for core-fucosylated N-glycan recognition.
Int.J.Biol.Macromol., 255, 2023
|
|
6JK3
 
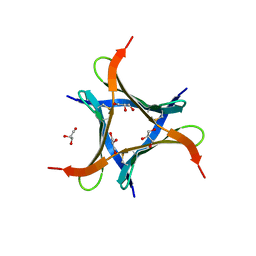 | | Crystal structure of a mini fungal lectin, PhoSL in complex with core-fucosylated chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lectin | | Authors: | Lou, Y.C, Chou, C.C, Yeh, H.H, Chien, C.Y, Sushant, S, Chen, C, Hsu, C.H. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the role of N-terminal integrity in PhoSL for core-fucosylated N-glycan recognition.
Int.J.Biol.Macromol., 255, 2023
|
|
6IDO
 
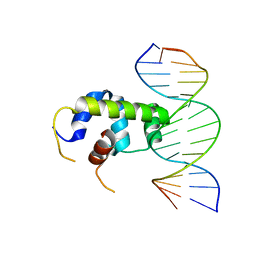 | | Crystal structure of Klebsiella pneumoniae sigma4 of sigmaS fusing with the RNA polymerase beta-flap-tip-helix in complex with -35 element DNA | | Descriptor: | DNA (5'-D(P*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*AP*AP*TP*CP*G)-3'), DNA (5'-D(P*GP*AP*TP*TP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*C)-3'), RNA polymerase sigma factor RpoS,RNA polymerase beta-flap-tip-helix | | Authors: | Lou, Y.C, Chien, C.Y, Chen, C, Hsu, C.H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.748 Å) | | Cite: | Structural basis for -35 element recognition by sigma4chimera proteins and their interactions with PmrA response regulator.
Proteins, 88, 2020
|
|
2KDZ
 
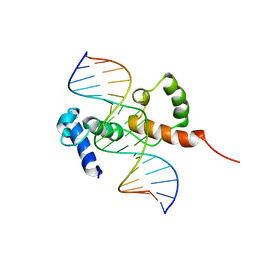 | | Structure of the R2R3 DNA binding domain of MYB1 protein from protozoan parasite trichomonas vaginalis in complex with MRE-1/MRE-2R DNA | | Descriptor: | 5'-D(*AP*AP*GP*AP*TP*AP*AP*CP*GP*AP*TP*AP*TP*TP*TP*A)-3', 5'-D(*TP*AP*AP*AP*TP*AP*TP*CP*GP*TP*TP*AP*TP*CP*TP*T)-3', MYB24 | | Authors: | Lou, Y.C, Wei, S.Y, Rajasekaran, M, Chou, C.C, Hsu, H.M, Tai, J.H, Chen, C. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of DNA recognition by a novel Myb1 DNA-binding domain in the protozoan parasite Trichomonas vaginalis.
Nucleic Acids Res., 2009
|
|
1BY0
 
 | |
2M87
 
 | |
2KXT
 
 | |
2KXV
 
 | |
1PJW
 
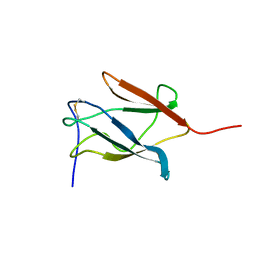 | | Solution Structure of the Domain III of the Japan Encephalitis Virus Envelope Protein | | Descriptor: | envelope protein | | Authors: | Wu, K.P, Wu, C.W, Tsao, Y.P, Lou, Y.C, Lin, C.W. | | Deposit date: | 2003-06-04 | | Release date: | 2003-11-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of a Flavivirus Recognized by Its Neutralizing Antibody: SOLUTION STRUCTURE OF THE DOMAIN III OF THE JAPANESE ENCEPHALITIS VIRUS ENVELOPE PROTEIN.
J.Biol.Chem., 278, 2003
|
|
1QLY
 
 | | NMR Study of the SH3 Domain From Bruton's Tyrosine Kinase, 20 Structures | | Descriptor: | TYROSINE-PROTEIN KINASE BTK | | Authors: | Tzeng, S.R, Lou, Y.C, Pai, M.T, Chen, C, Chen, S.H, Cheng, J.Y. | | Deposit date: | 1999-09-20 | | Release date: | 1999-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Human Btk SH3 Domain Complexed with a Proline-Rich Peptide from P120Cbl
J.Biomol.NMR, 16, 2000
|
|
1X37
 
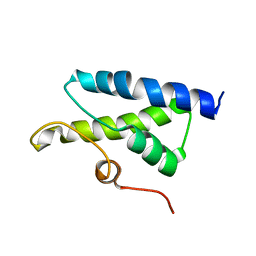 | | Structure of Bacillus subtilis Lon protease SSD domain | | Descriptor: | ATP-dependent protease La 1 | | Authors: | Wang, I, Lou, Y.C, Lo, S.C, Lee, Y.L, Wu, S.H, Chen, C. | | Deposit date: | 2005-04-30 | | Release date: | 2005-10-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis and DNA binding property of SSD domain of Bacillus subtilis Lon protease
to be published
|
|
1XHH
 
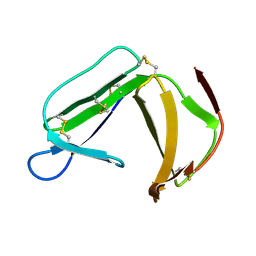 | | Solution Structure of porcine beta-microseminoprotein | | Descriptor: | beta-microseminoprotein | | Authors: | Wang, I, Lou, Y.C, Wu, K.P, Wu, S.H, Chang, W.C, Chen, C. | | Deposit date: | 2004-09-20 | | Release date: | 2005-03-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Novel solution structure of porcine beta-microseminoprotein
J.Mol.Biol., 346, 2005
|
|
2GO0
 
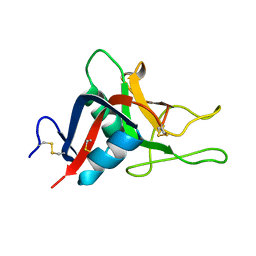 | |
