5CKD
 
 | | E. coli MazF E24A form III | | Descriptor: | Endoribonuclease MazF, SODIUM ION | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CO7
 
 | | E. coli MazF form III | | Descriptor: | Endoribonuclease MazF | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-19 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.489 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
3DIE
 
 | |
5CK9
 
 | | E. coli MazF form I | | Descriptor: | Endoribonuclease MazF, PENTAETHYLENE GLYCOL | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CKH
 
 | | E. coli MazF E24A form IIb | | Descriptor: | Endoribonuclease MazF | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CQX
 
 | |
5CW7
 
 | |
1LOY
 
 | | X-ray structure of the H40A/E58A mutant of Ribonuclease T1 complexed with 3'-guanosine monophosphate | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1 | | Authors: | Mignon, P, Steyaert, J, Loris, R, Geerlings, P, Loverix, S. | | Deposit date: | 2002-05-07 | | Release date: | 2002-08-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A nucleophile activation dyad in ribonucleases. A combined X-ray crystallographic/ab initio quantum chemical study
J.Biol.Chem., 277, 2002
|
|
1LOW
 
 | | X-ray structure of the H40A mutant of Ribonuclease T1 complexed with 3'-guanosine monophosphate | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1 | | Authors: | Mignon, P, Steyaert, J, Loris, R, Geerlings, P, Loverix, S. | | Deposit date: | 2002-05-07 | | Release date: | 2002-08-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A nucleophile activation dyad in ribonucleases. A combined X-ray crystallographic/ab initio quantum chemical study
J.Biol.Chem., 277, 2002
|
|
3G7Z
 
 | | CcdB dimer in complex with two C-terminal CcdA domains | | Descriptor: | Cytotoxic protein ccdB, Protein ccdA | | Authors: | De Jonge, N, Loris, R, Garcia-Pino, A, Buts, L. | | Deposit date: | 2009-02-11 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Rejuvenation of CcdB-Poisoned Gyrase by an Intrinsically Disordered Protein Domain.
Mol.Cell, 35, 2009
|
|
5CR2
 
 | |
5CQY
 
 | |
3DWT
 
 | |
1LU2
 
 | | DOLICHOS BIFLORUS SEED LECTIN IN COMPLEX WITH THE BLOOD GROUP A TRISACCHARIDE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, LECTIN, ... | | Authors: | Hamelryck, T.W, Loris, R, Bouckaert, J, Strecker, G, Imberty, A, Fernandez, E, Wyns, L, Etzler, M.E. | | Deposit date: | 1998-07-30 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
1LU1
 
 | | THE STRUCTURE OF THE DOLICHOS BIFLORUS SEED LECTIN IN COMPLEX WITH THE FORSSMAN DISACCHARIDE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ADENINE, CALCIUM ION, ... | | Authors: | Hamelryck, T.W, Loris, R, Bouckaert, J, Strecker, G, Imberty, A, Fernandez, E, Wyns, L, Etzler, M.E. | | Deposit date: | 1998-07-24 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
1LOV
 
 | | X-ray structure of the E58A mutant of Ribonuclease T1 complexed with 3'-guanosine monophosphate | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1 | | Authors: | Mignon, P, Steyaert, J, Loris, R, Geerlings, P, Loverix, S. | | Deposit date: | 2002-05-07 | | Release date: | 2002-08-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A nucleophile activation dyad in ribonucleases. A combined X-ray crystallographic/ab initio quantum chemical study
J.Biol.Chem., 277, 2002
|
|
5DLO
 
 | | S. aureus MazF in complex with substrate analogue | | Descriptor: | CHLORIDE ION, DNA substrate analogue AUACAUA, Endoribonuclease MazF | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-09-07 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Substrate recognition, regulation mechanism and activity regulation of MazF mRNA interferase.
To Be Published
|
|
3DD7
 
 | |
3DCQ
 
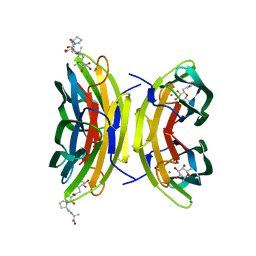 | | LECB (PA-LII) in complex with the synthetic ligand 2G0 | | Descriptor: | (2S)-1-[(2S)-6-amino-2-({[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyltetrahydro-2H-pyran-2-yl]acetyl}amino)hexanoyl]-N-[(1S)-1-carbamoyl-3-methylbutyl]pyrrolidine-2-carboxamide, CALCIUM ION, Fucose-binding lectin PA-IIL | | Authors: | Johansson, E.M, Crusz, S.A, Kolomiets, E, Buts, L, Kadam, R.U, Cacciarini, M, Bartels, K.M, Diggle, S.P, Camara, M, Williams, P, Loris, R, Nativi, C, Rosenau, F, Jaeger, K.E, Darbre, T, Reymond, J.L. | | Deposit date: | 2008-06-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition and dispersion of Pseudomonas aeruginosa biofilms by glycopeptide dendrimers targeting the fucose-specific lectin LecB.
Chem.Biol., 15, 2008
|
|
1G8W
 
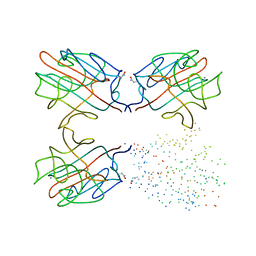 | | IMPROVED STRUCTURE OF PHYTOHEMAGGLUTININ-L FROM THE KIDNEY BEAN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LEUCOAGGLUTINATING PHYTOHEMAGGLUTININ, ... | | Authors: | Buts, L, Hamelryck, T.W, Dao-Thi, M, Loris, R, Wyns, L, Etzler, M.E. | | Deposit date: | 2000-11-21 | | Release date: | 2000-12-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Weak protein-protein interactions in lectins: the crystal structure of a vegetative lectin from the legume Dolichos biflorus.
J.Mol.Biol., 309, 2001
|
|
3ENR
 
 | | ZINC-CALCIUM CONCANAVALIN A AT PH 6.15 | | Descriptor: | CALCIUM ION, Concanavalin-A, ZINC ION | | Authors: | Bouckaert, J, Loris, R, Wyns, L. | | Deposit date: | 1999-02-11 | | Release date: | 2000-09-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Zinc/calcium- and cadmium/cadmium-substituted concanavalin A: interplay of metal binding, pH and molecular packing.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1ENQ
 
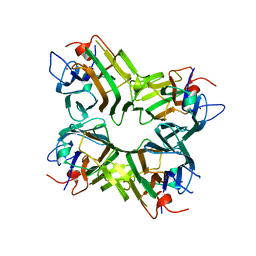 | | CO-CRYSTALS OF DEMETALLIZED CONCANAVALIN A WITH ZINC HAVING A ZINC ION BOUND IN THE S1 SITE | | Descriptor: | CONCANAVALIN A, ZINC ION | | Authors: | Bouckaert, J, Loris, R, Poortmans, F, Wyns, L. | | Deposit date: | 1996-03-20 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequential structural changes upon zinc and calcium binding to metal-free concanavalin A.
J.Biol.Chem., 271, 1996
|
|
1APN
 
 | |
1BVI
 
 | | RIBONUCLEASE T1 (WILDTYPE) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-15 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
1DBN
 
 | | MAACKIA AMURENSIS LEUKOAGGLUTININ (LECTIN) WITH SIALYLLACTOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Imberty, A, Gautier, C, Lescar, J, Loris, R. | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | An unusual carbohydrate binding site revealed by the structures of two Maackia amurensis lectins complexed with sialic acid-containing oligosaccharides.
J.Biol.Chem., 275, 2000
|
|
