5HTE
 
 | | Recombinant bovine beta-lactoglobulin variant L1A/I2S (sBlgB#2) | | Descriptor: | Beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Tworzydlo, M, Polit, A, Hawro, B, Lach, A, Ludwin, E, Lewinski, K. | | Deposit date: | 2016-01-26 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Engineered beta-Lactoglobulin Produced in E. coli: Purification, Biophysical and Structural Characterisation.
Mol Biotechnol., 58, 2016
|
|
5HTD
 
 | | Recombinant bovine beta-lactoglobulin variant L1A/I2S with endogenous ligand (sBlgB#1) | | Descriptor: | Beta-lactoglobulin, MYRISTIC ACID | | Authors: | Loch, J.I, Bonarek, P, Tworzydlo, M, Polit, A, Hawro, B, Lach, A, Ludwin, E, Lewinski, K. | | Deposit date: | 2016-01-26 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineered beta-Lactoglobulin Produced in E. coli: Purification, Biophysical and Structural Characterisation.
Mol Biotechnol., 58, 2016
|
|
6QI6
 
 | | Trigonal form of WT recombinant bovine beta-lactoglobulin | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactoglobulin, ETHANOL | | Authors: | Loch, J.I, Krawczyk, A, Lewinski, K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design approach to rational site-directed mutagenesis of beta-lactoglobulin.
J.Struct.Biol., 210, 2020
|
|
6QI7
 
 | | Engineered beta-lactoglobulin: variant L39Y in complex with endogenous ligand | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactoglobulin, PALMITIC ACID | | Authors: | Loch, J.I, Siuda, M.K, Lewinski, K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design approach to rational site-directed mutagenesis of beta-lactoglobulin.
J.Struct.Biol., 210, 2020
|
|
7R1G
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-38 (R207C, D210S, S211V) | | Descriptor: | Beta-aspartyl-peptidase, Isoaspartyl peptidase, SODIUM ION | | Authors: | Loch, J.I, Kadziolka, K, Jaskolski, M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6T44
 
 | |
6T42
 
 | |
6HCE
 
 | |
8OSW
 
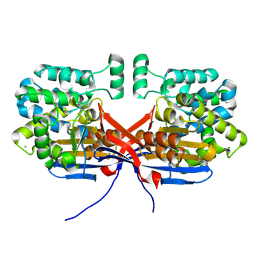 | | Crystal structure of Rhizobium etli L-asparaginase ReAIV (R4mC-1) | | Descriptor: | CHLORIDE ION, Putative L-asparaginase II protein, ZINC ION | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8ORI
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAIV (orthorhombic) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6XVE
 
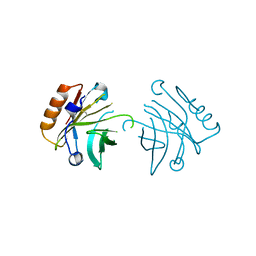 | |
7ZLF
 
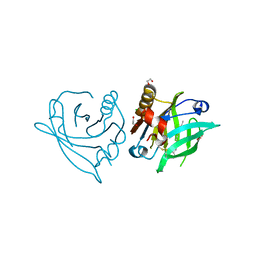 | | Mutant L39Y-L58F of recombinant bovine beta-lactoglobulin in complex with endogenous fatty acid | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactoglobulin, CHLORIDE ION, ... | | Authors: | Loch, J.I, Kurpiewska, K, Bonarek, P. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | beta-Lactoglobulin variants as potential carriers of pramoxine: Comprehensive structural and biophysical studies.
J.Mol.Recognit., 36, 2023
|
|
7ZCD
 
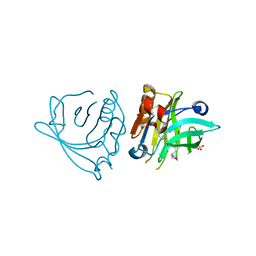 | |
7ZA0
 
 | |
7Z9Z
 
 | | Mutant L39Y of recombinant bovine beta-lactoglobulin in complex with pramocaine | | Descriptor: | Beta-lactoglobulin, CHLORIDE ION, Pramocaine, ... | | Authors: | Loch, J.I, Kurpiewska, K, Bonarek, P. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | beta-Lactoglobulin variants as potential carriers of pramoxine: Comprehensive structural and biophysical studies.
J.Mol.Recognit., 36, 2023
|
|
4Y0S
 
 | | Goat beta-lactoglobulin complex with pramocaine (GLG-PRM) | | Descriptor: | Beta-lactoglobulin, Pramocaine, SULFATE ION | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Jablonski, M, Czub, M, Ye, X, Lewinski, K. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | beta-Lactoglobulin interactions with local anaesthetic drugs - Crystallographic and calorimetric studies.
Int.J.Biol.Macromol., 80, 2015
|
|
4Y0Q
 
 | | Bovine beta-lactoglobulin complex with pramocaine crystallized from sodium citrate (BLG-PRM1) | | Descriptor: | Beta-lactoglobulin, Pramocaine | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Jablonski, M, Czub, M, Ye, X, Lewinski, K. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | beta-Lactoglobulin interactions with local anaesthetic drugs - Crystallographic and calorimetric studies.
Int.J.Biol.Macromol., 80, 2015
|
|
4Y0R
 
 | | Bovine beta-lactoglobulin complex with pramocaine crystallized from ammonium sulphate (BLG-PRM2) | | Descriptor: | Beta-lactoglobulin, Pramocaine | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Jablonski, M, Czub, M, Ye, X, Lewinski, K. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | beta-Lactoglobulin interactions with local anaesthetic drugs - Crystallographic and calorimetric studies.
Int.J.Biol.Macromol., 80, 2015
|
|
4Y0P
 
 | | Bovine beta-lactoglobulin complex with tetracaine (BLG-TET) | | Descriptor: | Beta-lactoglobulin, Tetracaine | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Jablonski, M, Czub, M, Ye, X, Lewinski, K. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | beta-Lactoglobulin interactions with local anaesthetic drugs - Crystallographic and calorimetric studies.
Int.J.Biol.Macromol., 80, 2015
|
|
5K06
 
 | | Recombinant bovine beta-lactoglobulin with uncleaved N-terminal methionine (rBlgB) | | Descriptor: | Beta-lactoglobulin, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Loch, J.I, Bonarek, P, Tworzydlo, M, Polit, A, Hawro, B, Lach, A, Ludwin, E, Lewinski, K. | | Deposit date: | 2016-05-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineered beta-Lactoglobulin Produced in E. coli: Purification, Biophysical and Structural Characterisation.
Mol Biotechnol., 58, 2016
|
|
7OS5
 
 | | Crystal structure of Rhizobium etli inducible L-asparaginase ReAV (orthorhombic form OP) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, L-asparaginase, ... | | Authors: | Loch, J.I, Imiolczyk, B, Gilski, M, Jaskolski, M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-11-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.293 Å) | | Cite: | Crystal structures of the elusive Rhizobium etli L-asparaginase reveal a peculiar active site.
Nat Commun, 12, 2021
|
|
7OS6
 
 | | Crystal structure of Rhizobium etli inducible L-asparaginase ReAV (monoclinic form MP1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Loch, J.I, Imiolczyk, B, Gilski, M, Jaskolski, M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-11-24 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structures of the elusive Rhizobium etli L-asparaginase reveal a peculiar active site.
Nat Commun, 12, 2021
|
|
6QPD
 
 | |
6QPE
 
 | |
5NUN
 
 | | Engineered beta-lactoglobulin: variant F105L-L39A-M107F in complex with chlorpromazine (LG-LAF-CLP) | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Towrzydlo, M, Lazinskia, I, Szydlowska, J, Lewinski, K. | | Deposit date: | 2017-04-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The engineered beta-lactoglobulin with complementarity to the chlorpromazine chiral conformers.
Int. J. Biol. Macromol., 114, 2018
|
|
